ID |
Date |
Author |
Subject |
|
33
|
Fri Aug 23 09:08:42 2024 |
AISHA | Weak s-process study (23/8/24) |
|
| Attachment 1: Friday-23_8.pptx
|
|
32
|
Sun Mar 17 12:11:37 2024 |
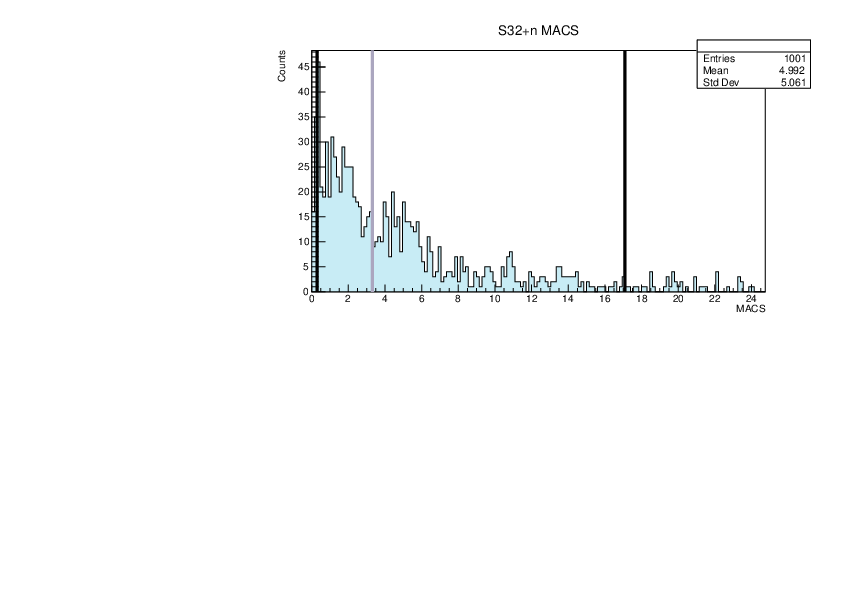
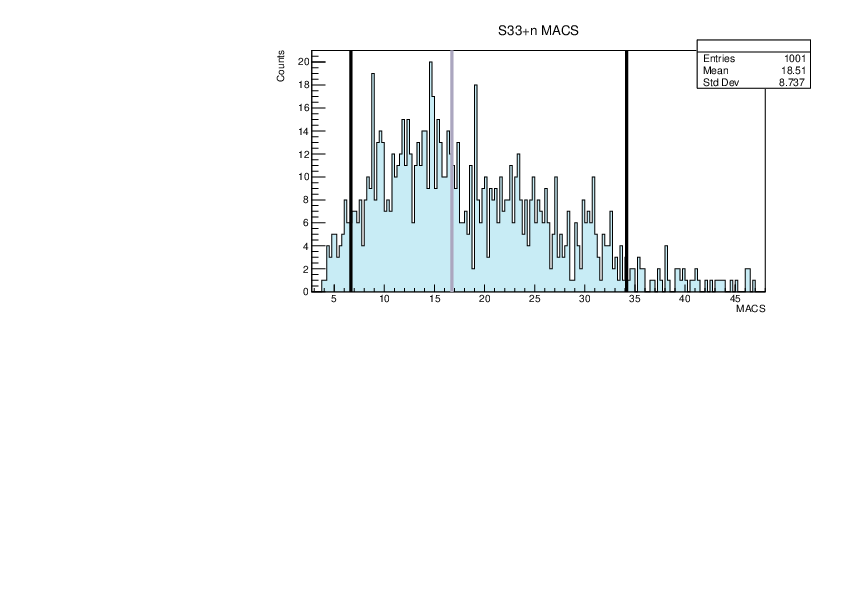
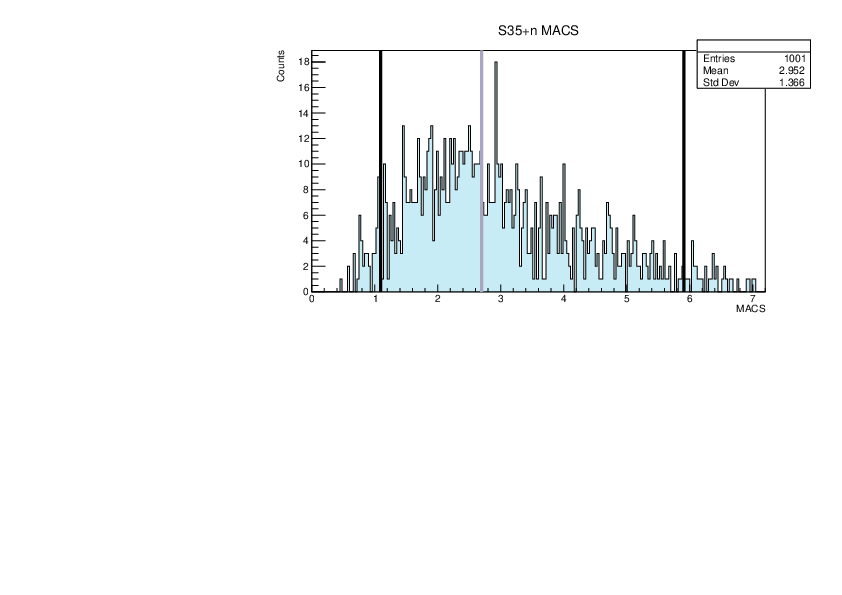
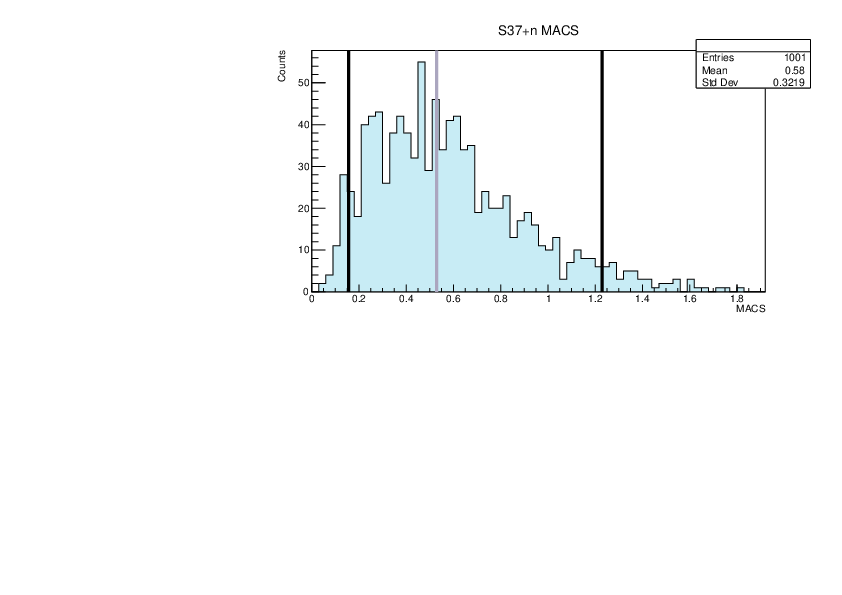
Selin Ezgi Birincioglu | myGOE.f Nuclei |
|
| Attachment 1: lim.pdf
|
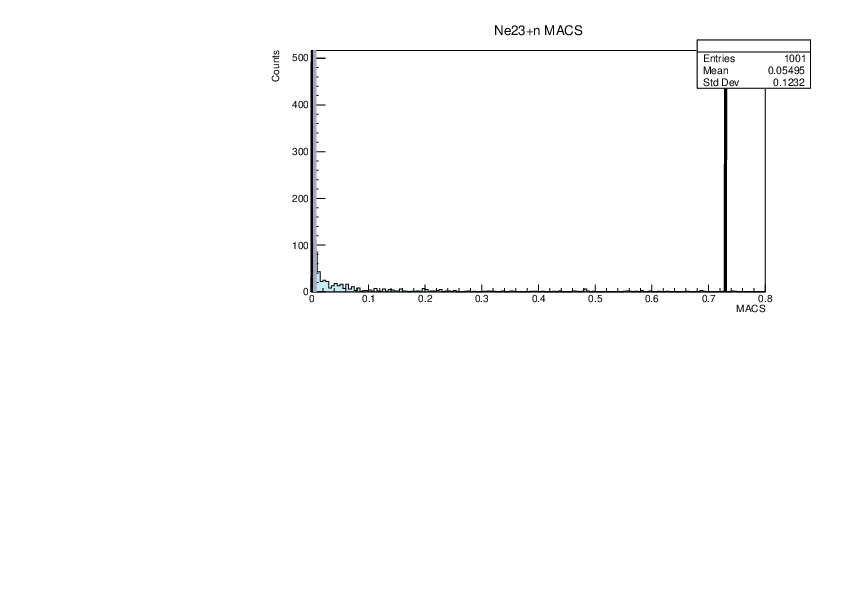
|
| Attachment 2: lim.pdf
|
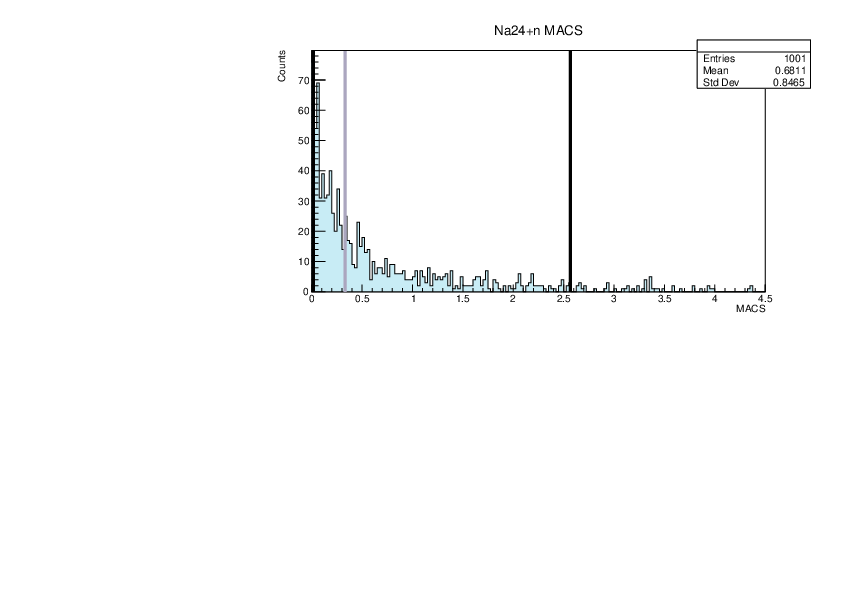
|
| Attachment 3: lim.pdf
|
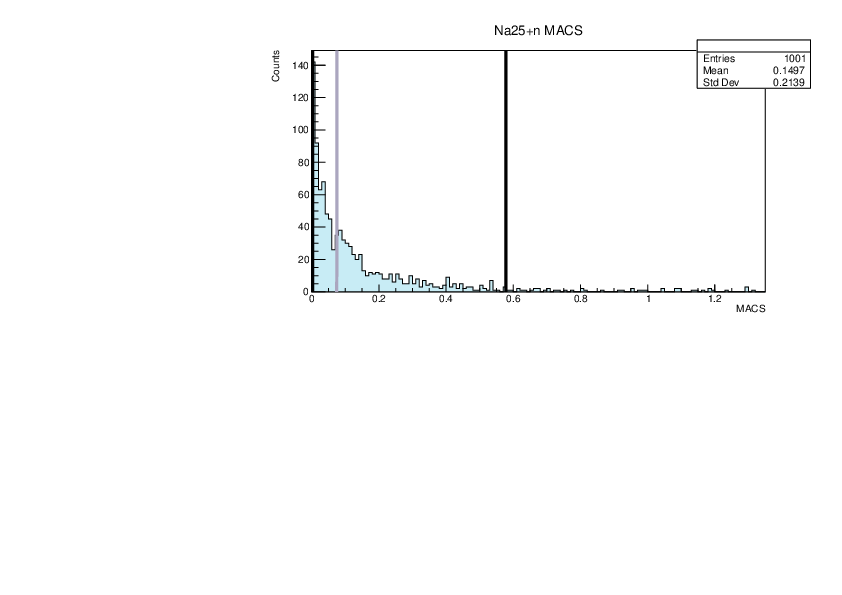
|
| Attachment 4: lim.pdf
|
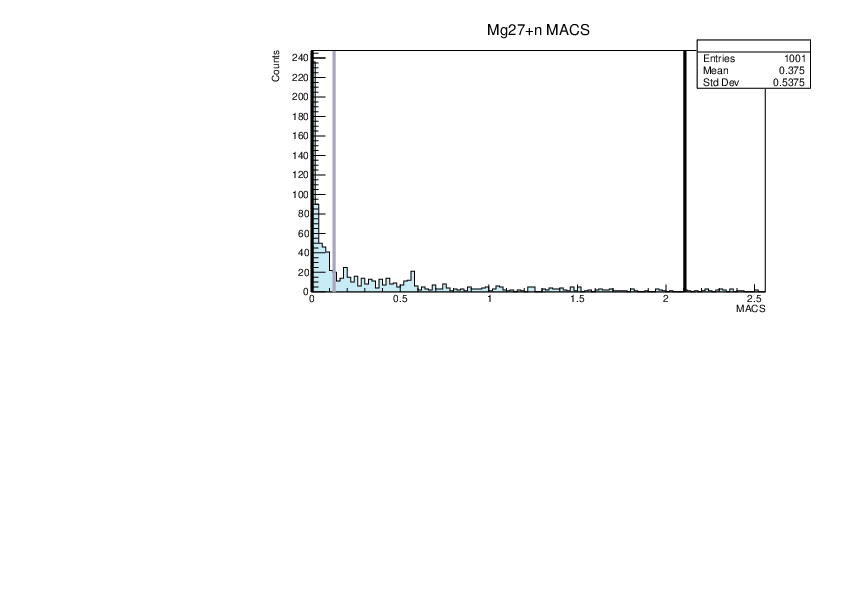
|
| Attachment 5: lim.pdf
|
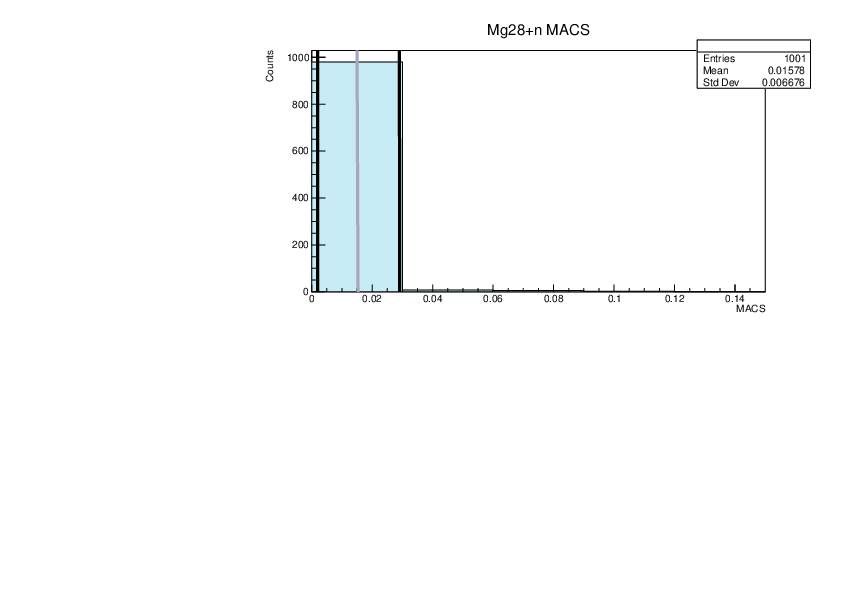
|
| Attachment 6: lim.pdf
|
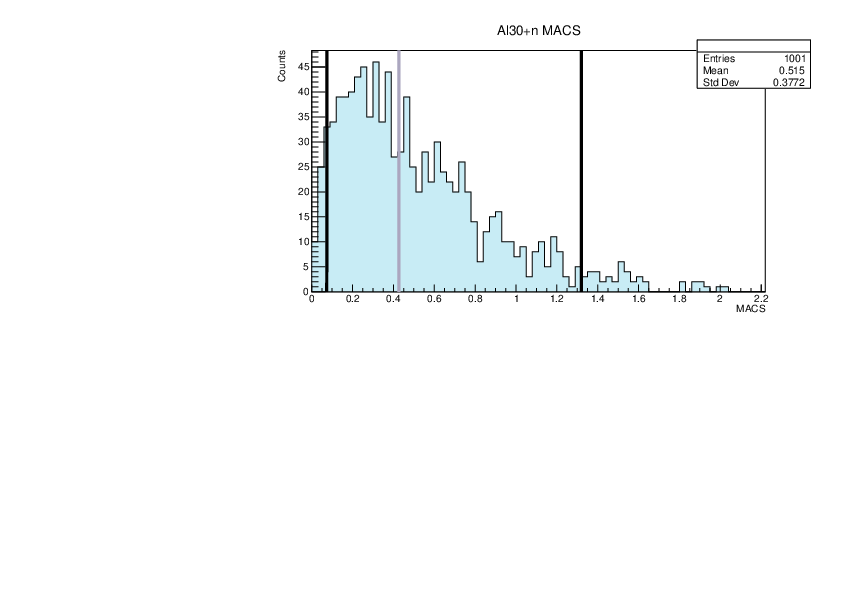
|
| Attachment 7: lim.pdf
|
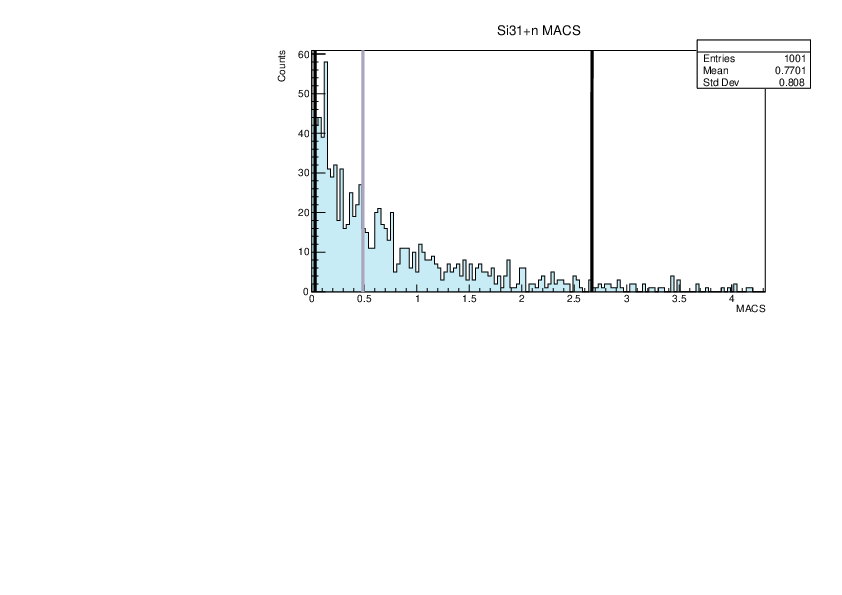
|
| Attachment 8: lim.pdf
|
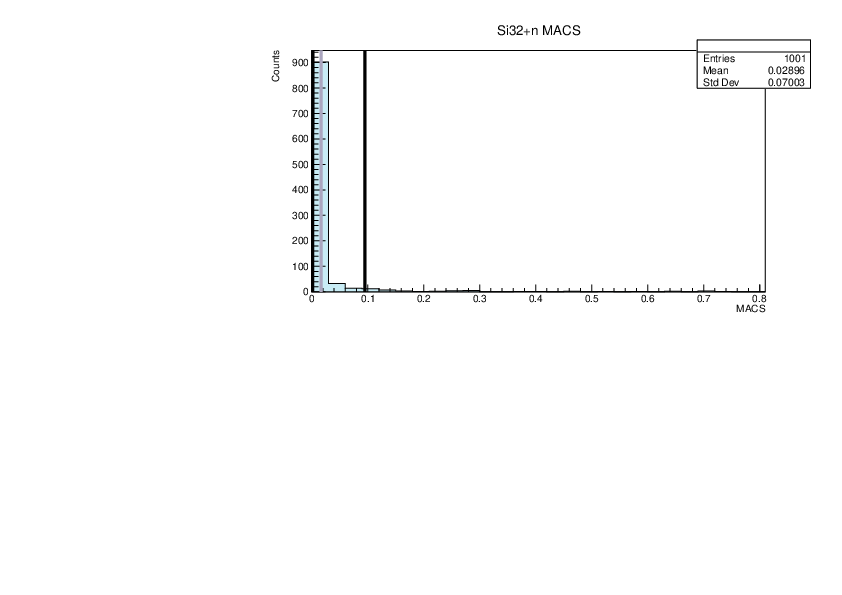
|
| Attachment 9: lim.pdf
|
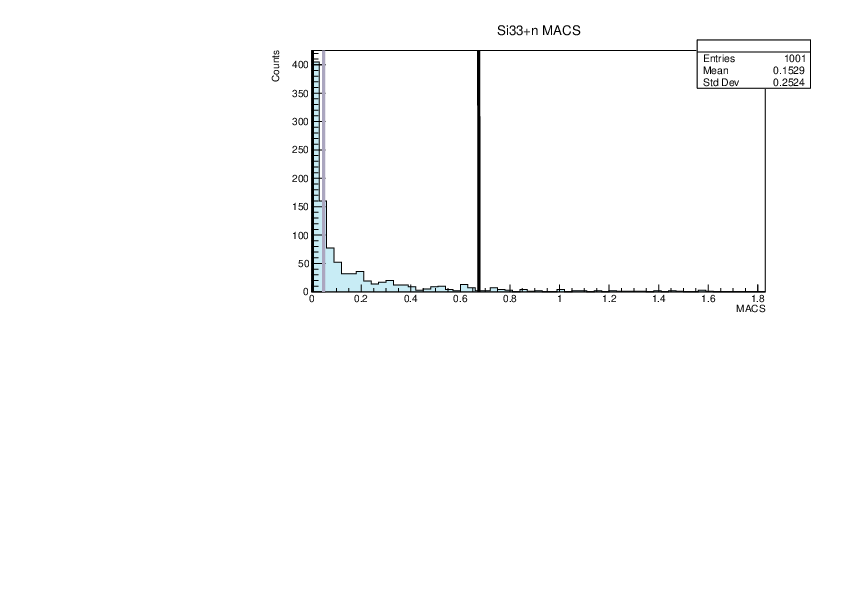
|
| Attachment 10: lim.pdf
|
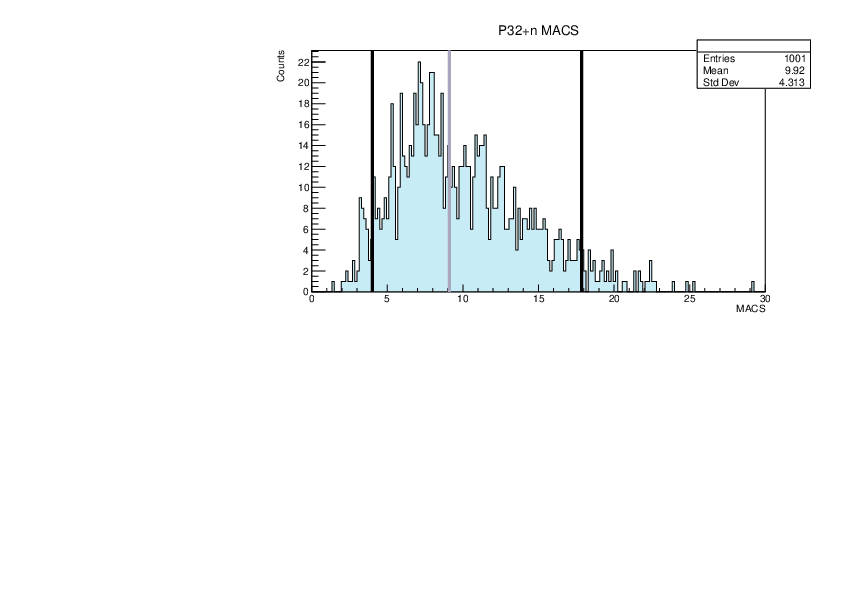
|
| Attachment 11: lim.pdf
|
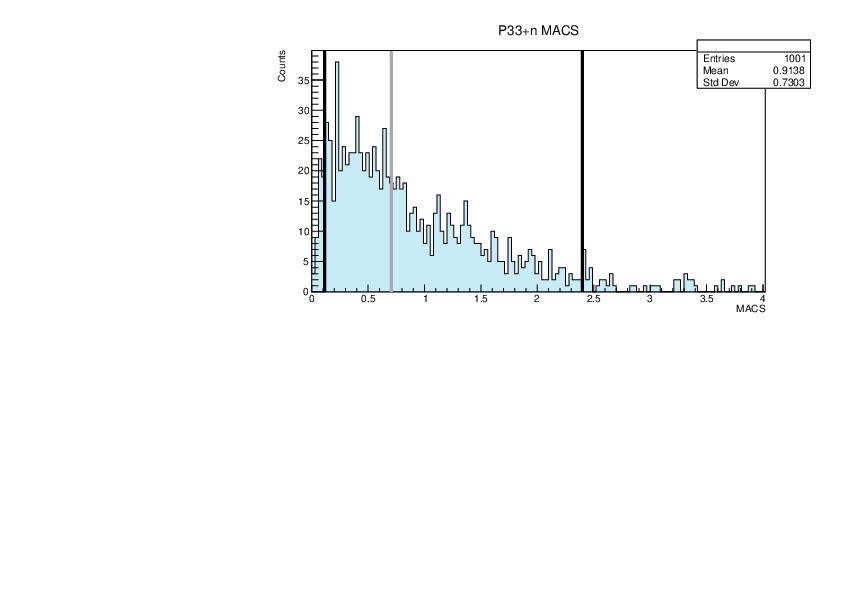
|
| Attachment 12: lim.pdf
|
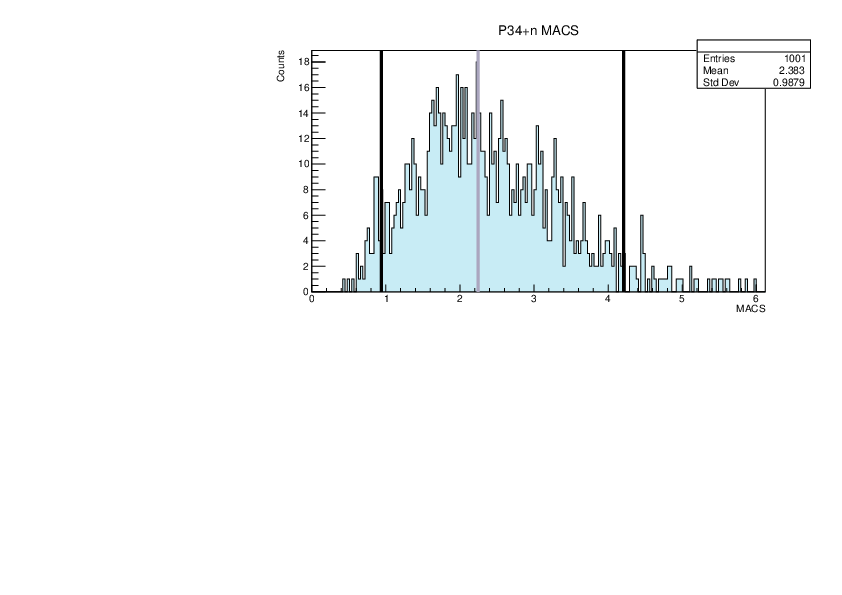
|
| Attachment 13: lim.pdf
|
|
| Attachment 14: lim.pdf
|
|
| Attachment 15: lim.pdf
|
|
| Attachment 16: lim.pdf
|
|
| Attachment 17: lim.pdf
|
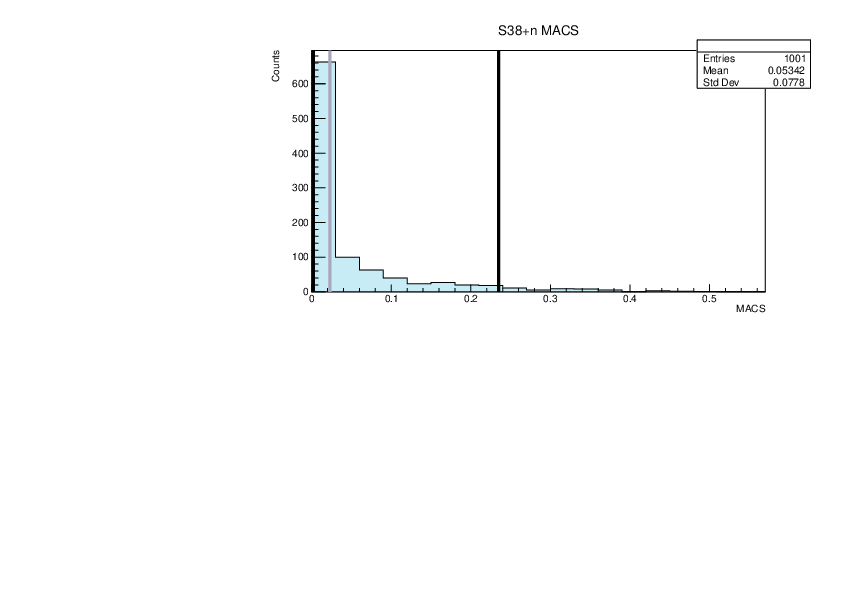
|
| Attachment 18: lim.pdf
|
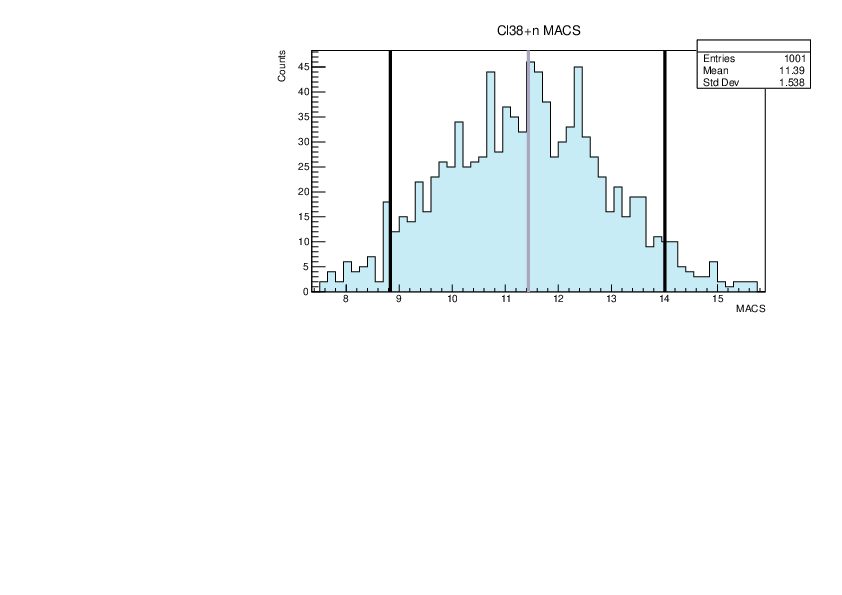
|
| Attachment 19: lim.pdf
|
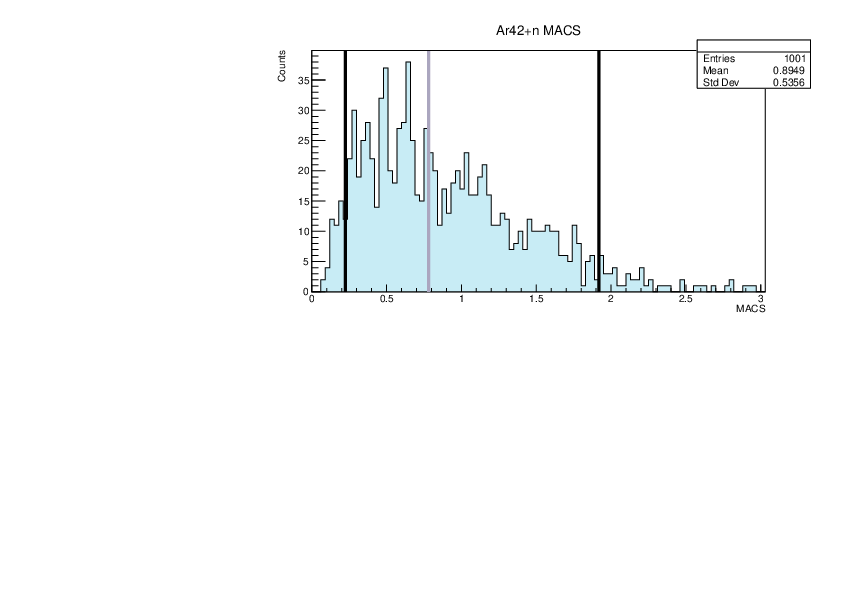
|
| Attachment 20: lim.pdf
|
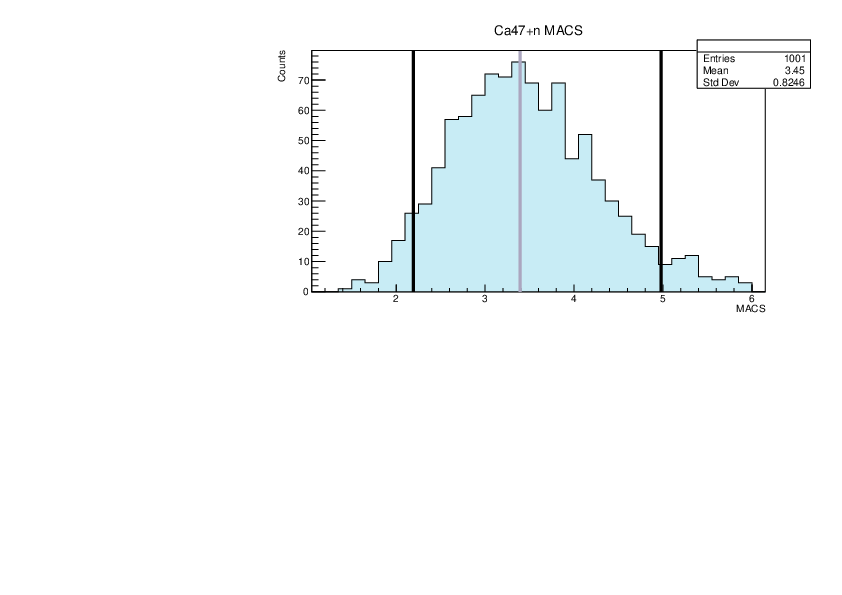
|
| Attachment 21: lim.pdf
|
|
|
31
|
Fri Mar 15 10:14:12 2024 |
Selin Ezgi Birincioglu | New MACS |
|
| Attachment 1: rates_clw.xlsx
|
|
30
|
Mon Mar 11 16:29:08 2024 |
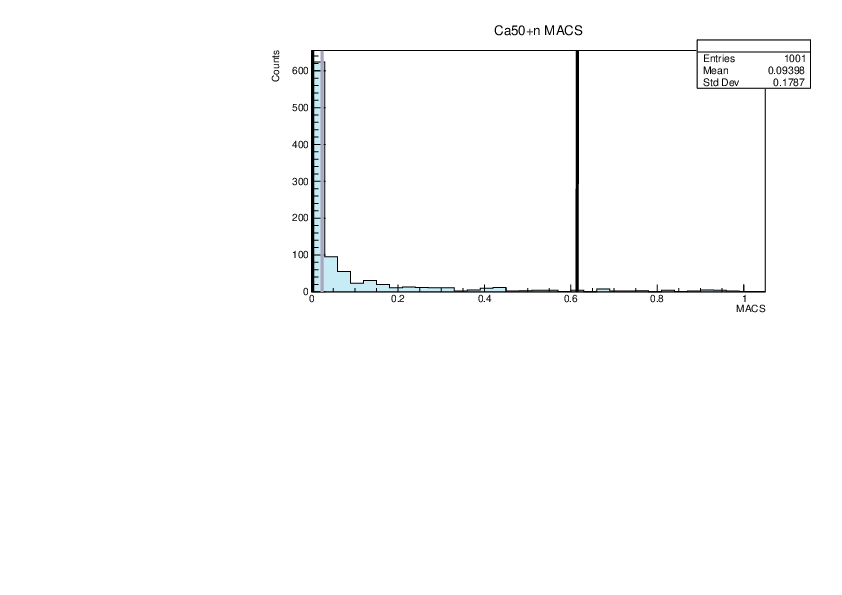
Selin Ezgi Birincioglu | Mixed Nuclei |
|
| Attachment 1: lim.pdf
|
|
| Attachment 2: lim.pdf
|
|
| Attachment 3: lim.pdf
|
|
| Attachment 4: lim.pdf
|
|
| Attachment 5: lim.pdf
|
|
| Attachment 6: lim.pdf
|
|
| Attachment 7: lim.pdf
|
|
| Attachment 8: lim.pdf
|
|
|
29
|
Mon Feb 19 21:42:39 2024 |
Selin Ezgi Birincioglu | MACS |
Excel Sheet of results, orange ones are nuclei that required both TALYS and mygoe.f. This file is colour coded - can be seen below the table. |
| Attachment 1: rates_clw.xlsx
|
|
28
|
Sat Feb 17 21:30:23 2024 |
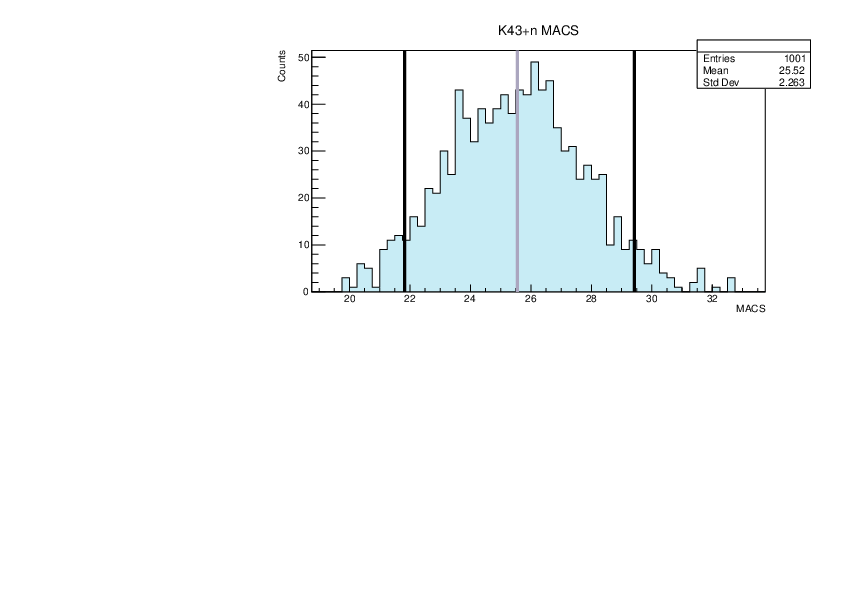
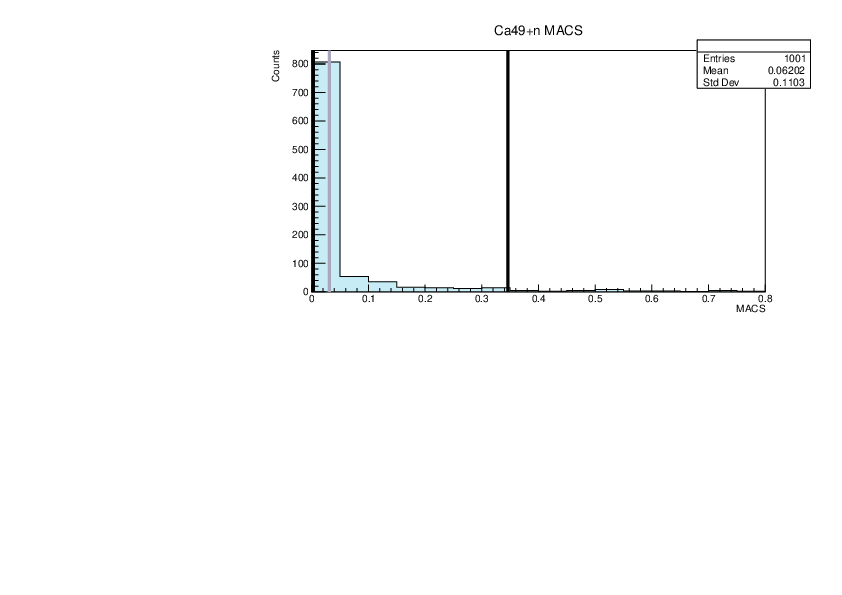
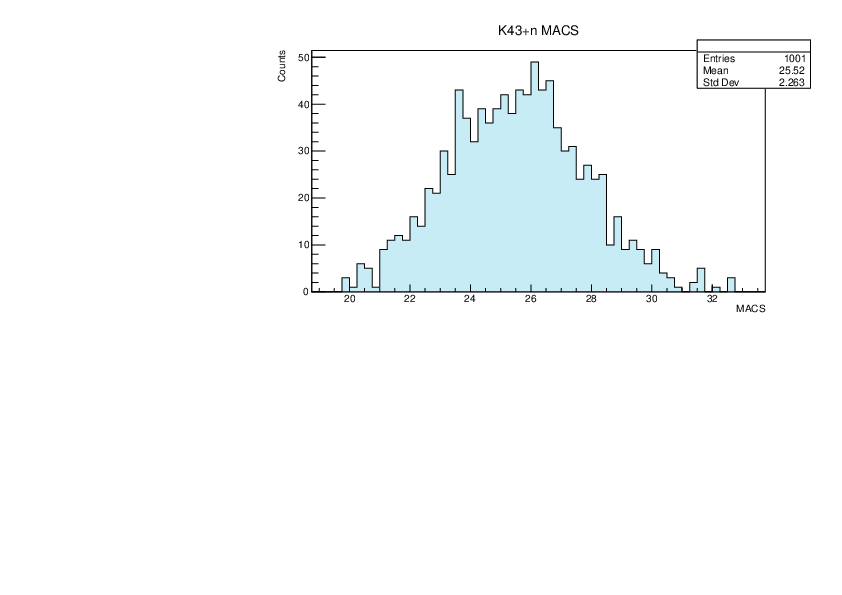
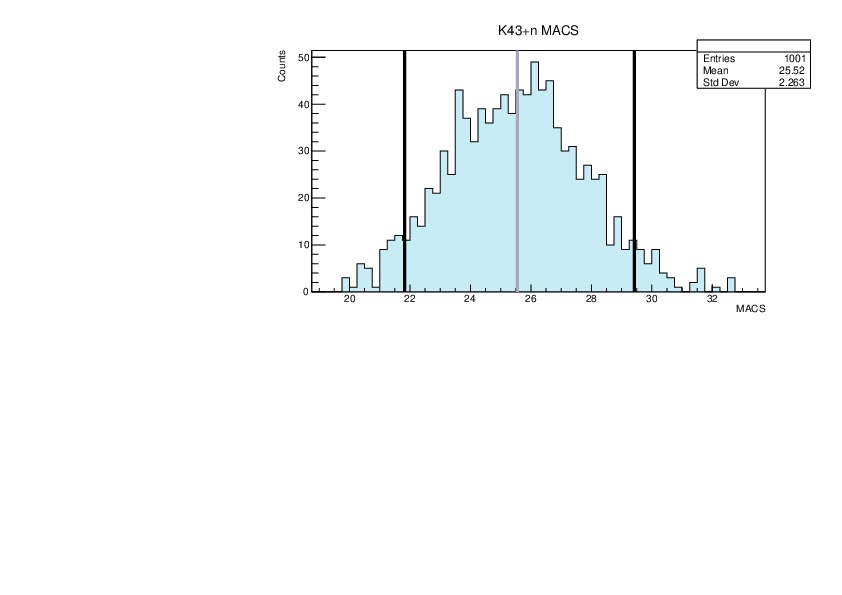
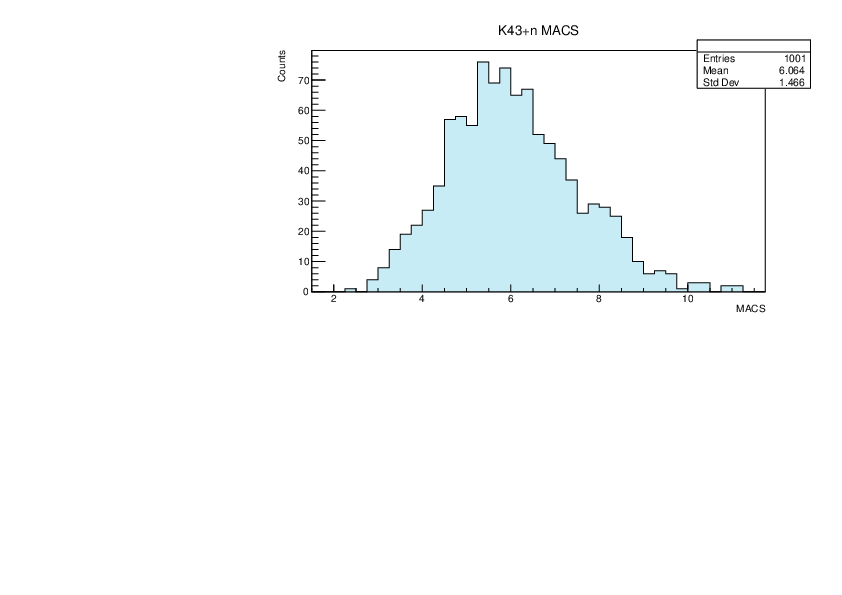
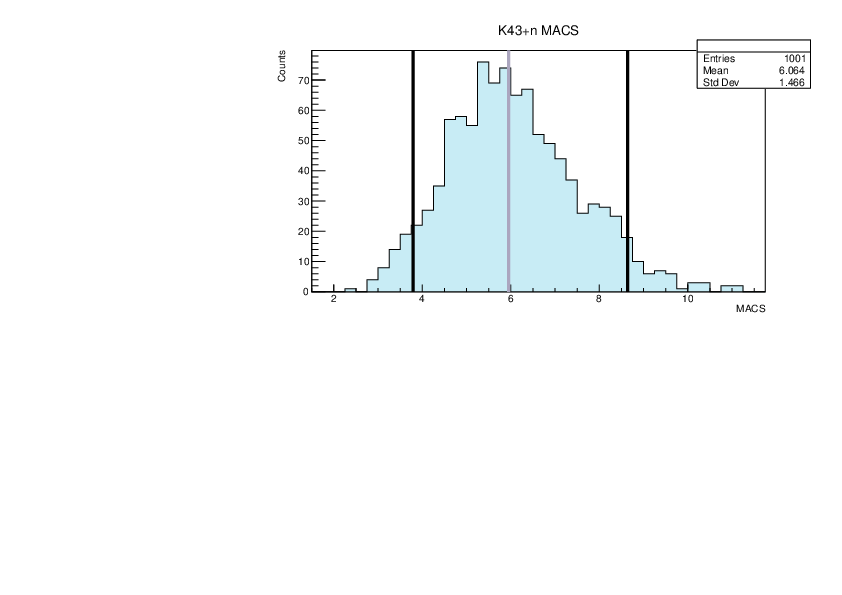
Selin Ezgi Birincioglu | K43+n Variations By Combination of Models |
K43-13 and -42. |
| Attachment 1: dirst.pdf
|
|
| Attachment 2: lim.pdf
|
|
| Attachment 3: dist.pdf
|
|
| Attachment 4: lim.pdf
|
|
|
27
|
Sat Feb 17 20:42:03 2024 |
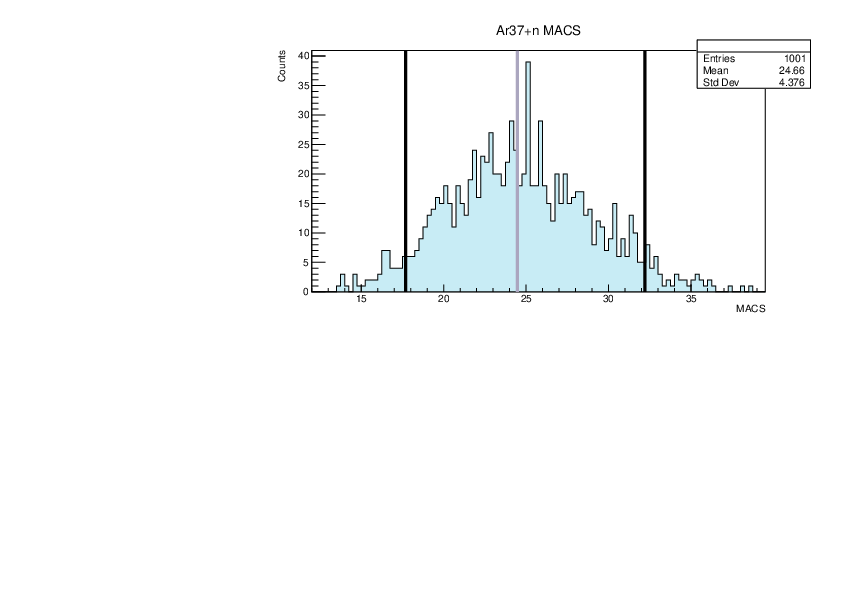
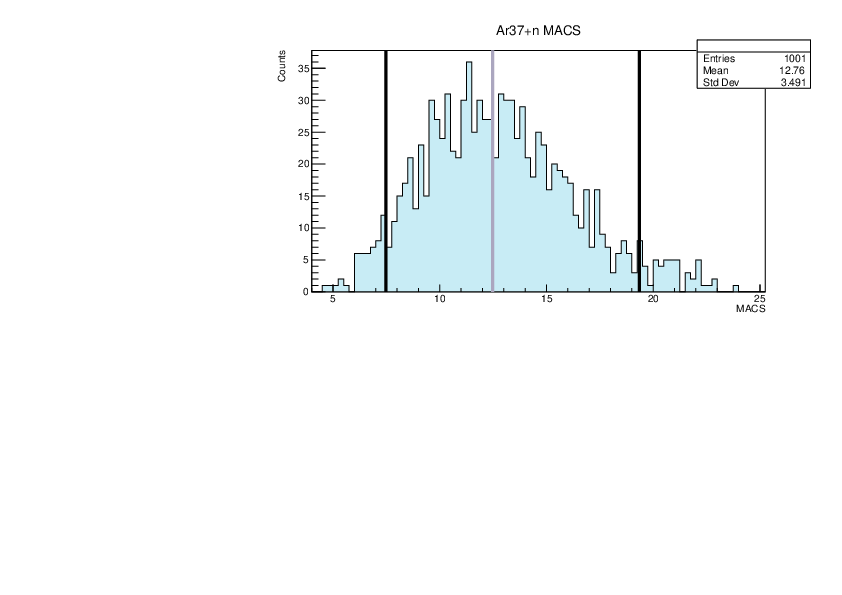
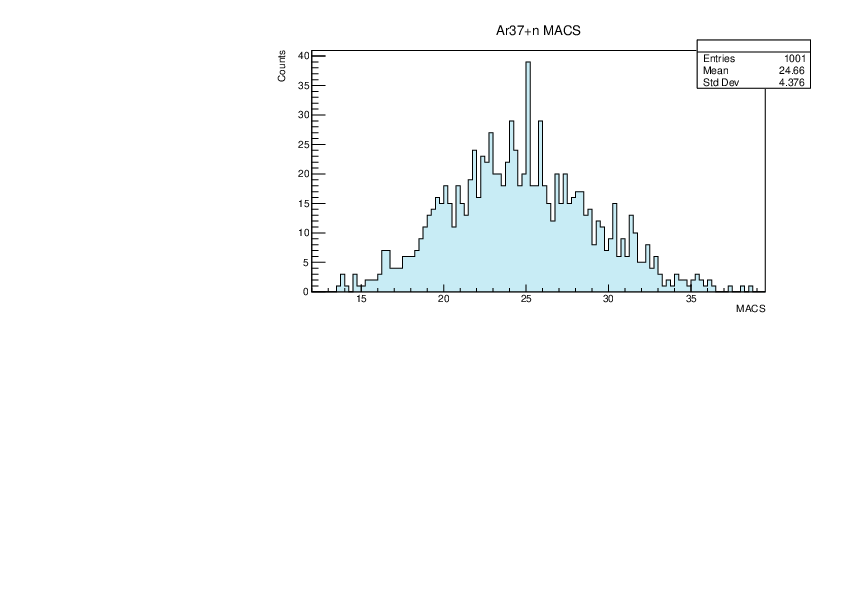
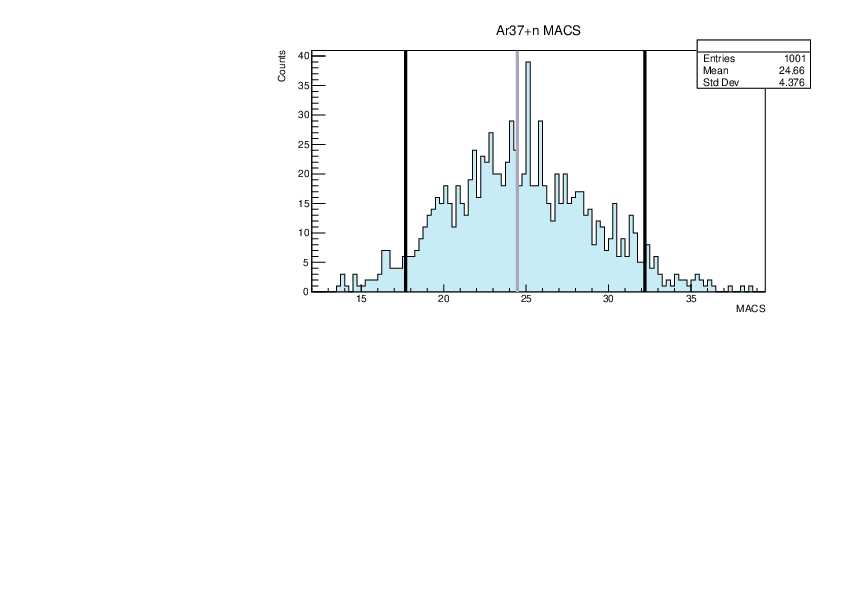
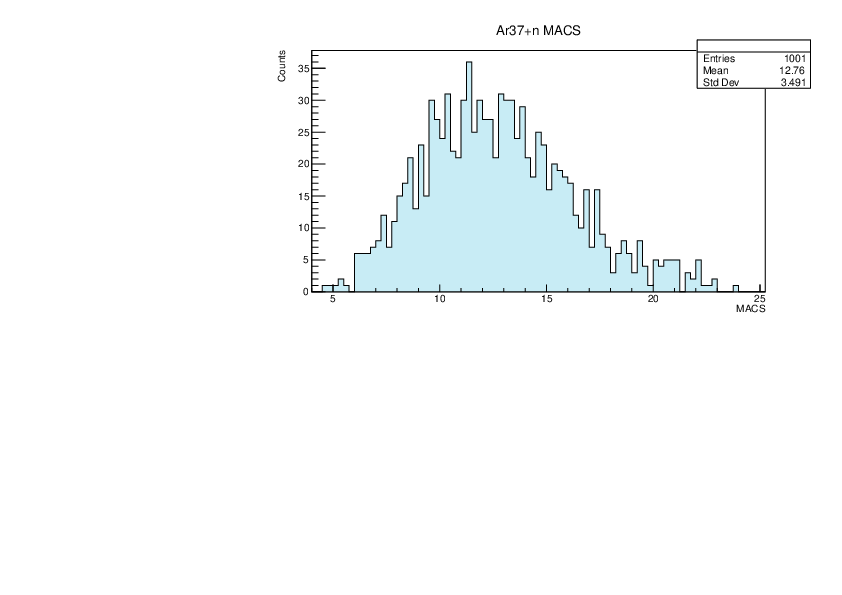
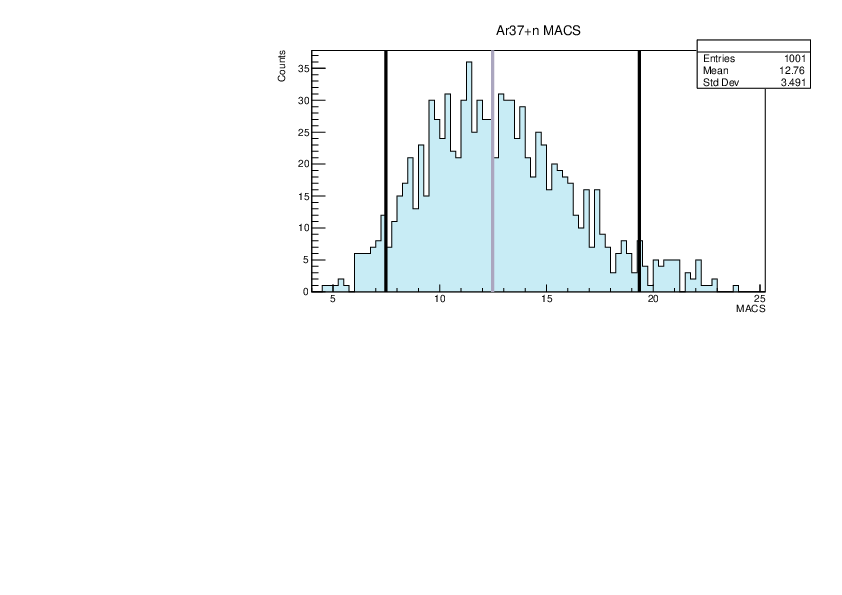
Selin Ezgi Birincioglu | Ar37+n Variations By Combination of Models |
Ar37-13 and -42. |
| Attachment 1: dist.pdf
|
|
| Attachment 2: lim.pdf
|
|
| Attachment 3: dist.pdf
|
|
| Attachment 4: lim.pdf
|
|
|
26
|
Sat Feb 17 20:35:21 2024 |
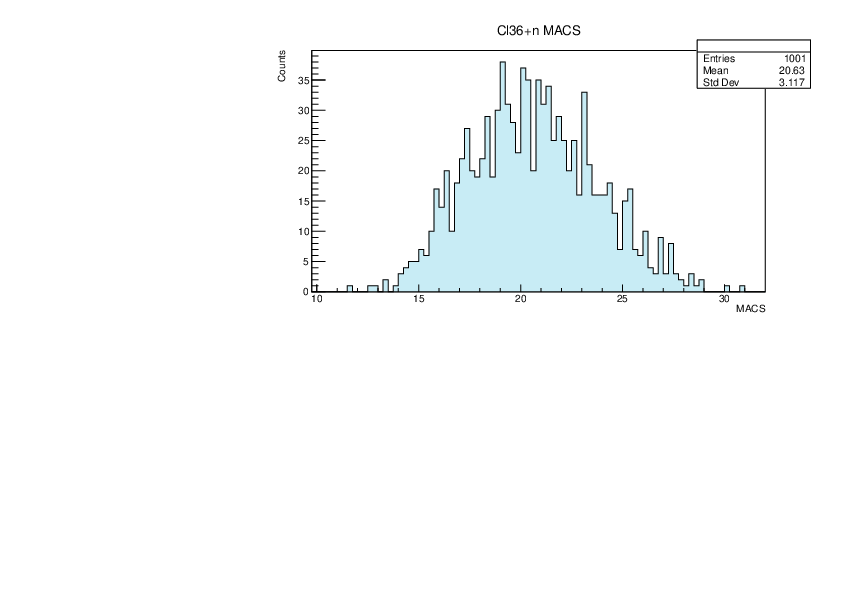
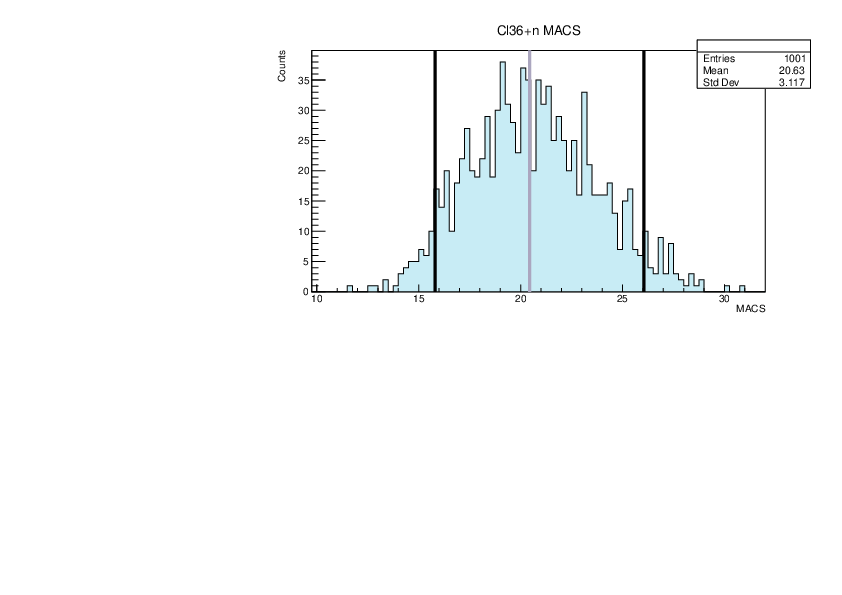
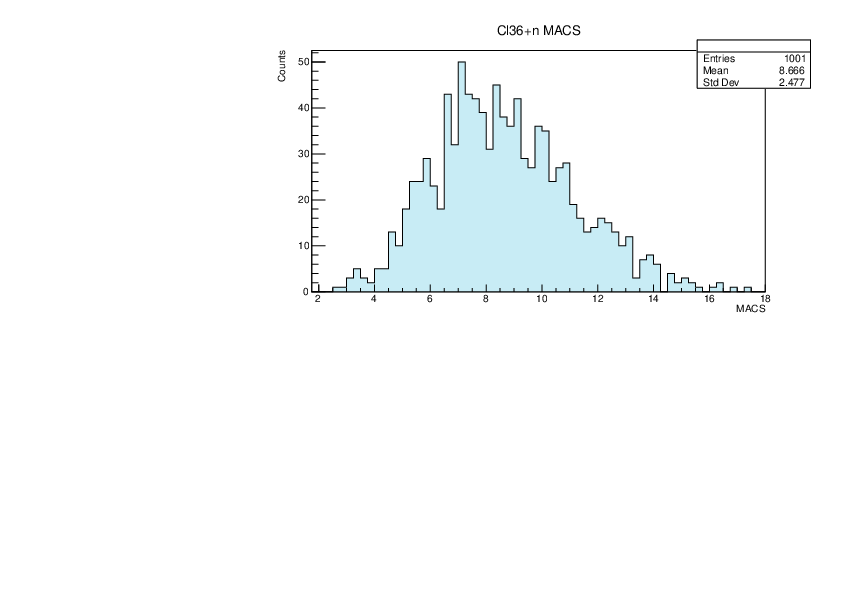
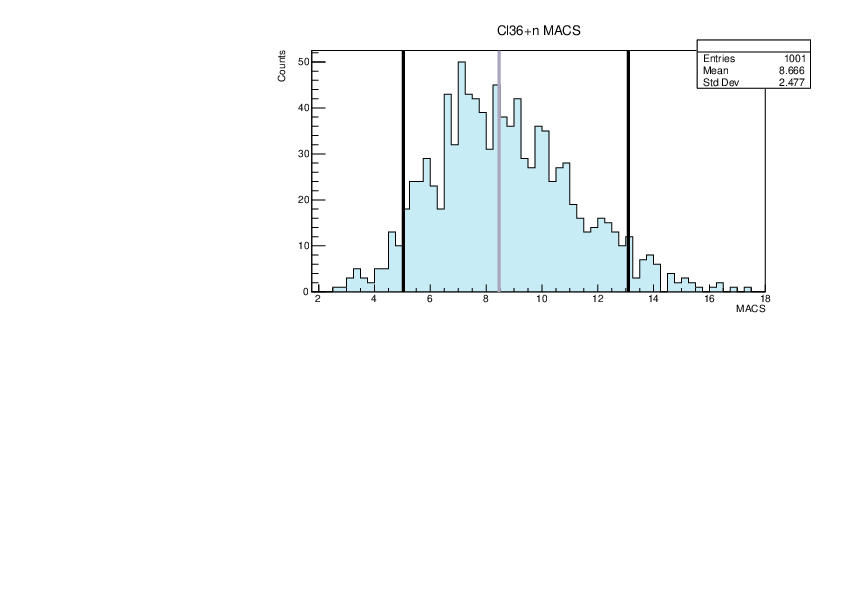
Selin Ezgi Birincioglu | Cl36+n Variations By Combination of Models |
Cl36-10 and -42. |
| Attachment 1: dist.pdf
|
|
| Attachment 2: lim.pdf
|
|
| Attachment 3: dist.pdf
|
|
| Attachment 4: lim.pdf
|
|
|
25
|
Sat Feb 17 20:18:15 2024 |
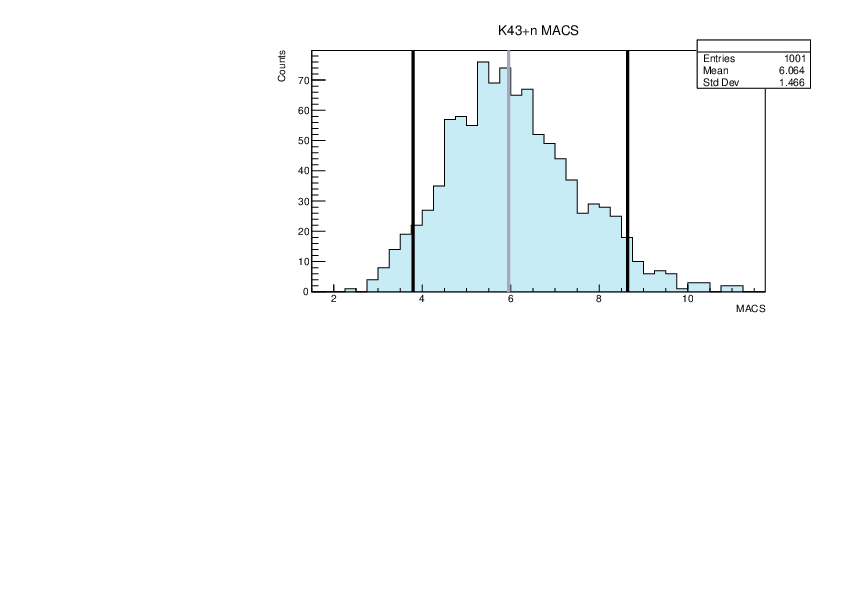
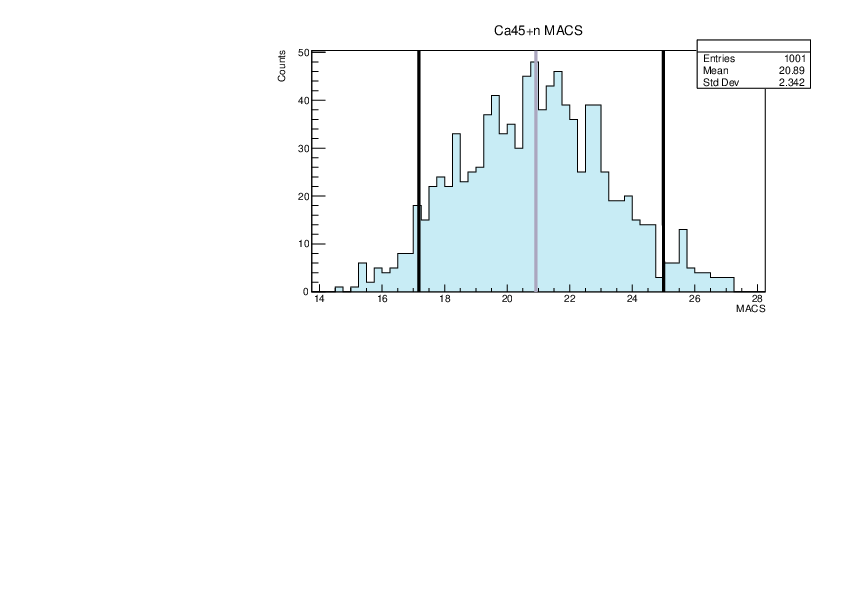
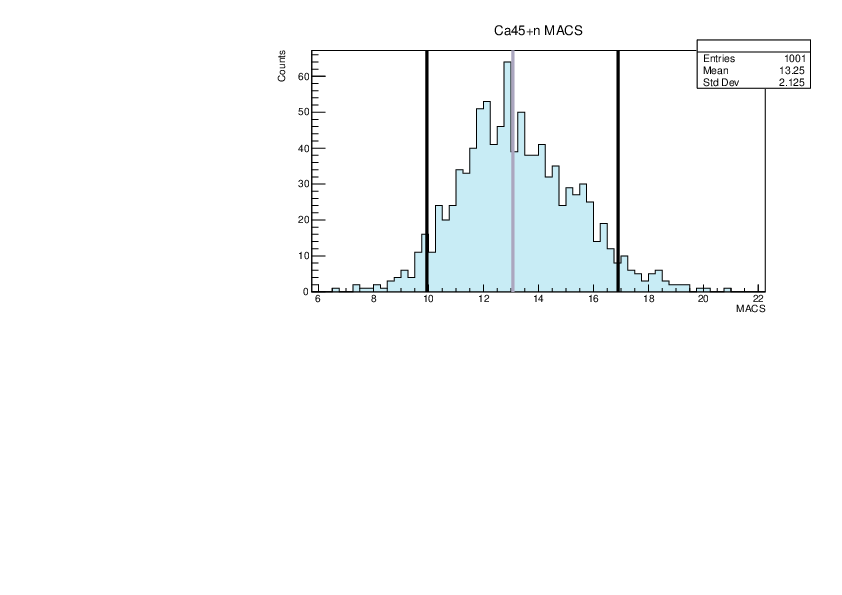
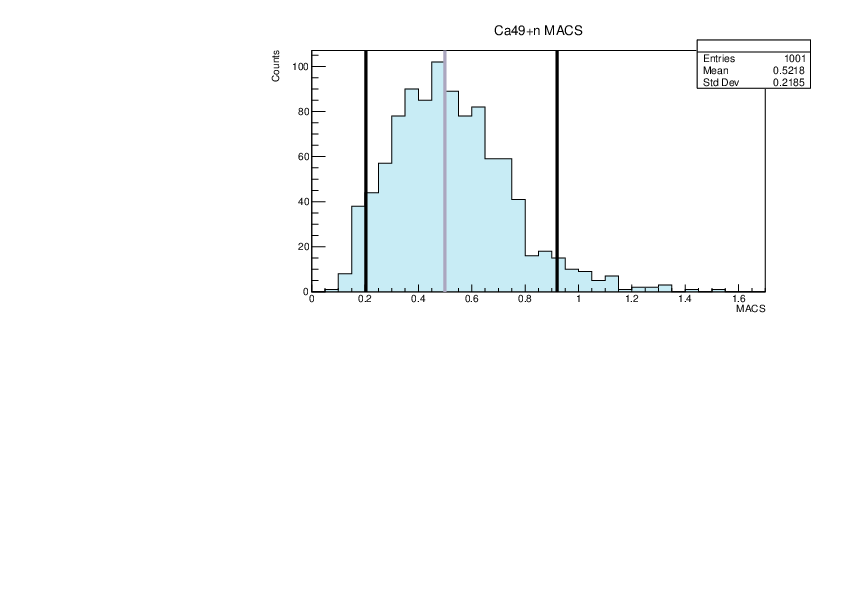
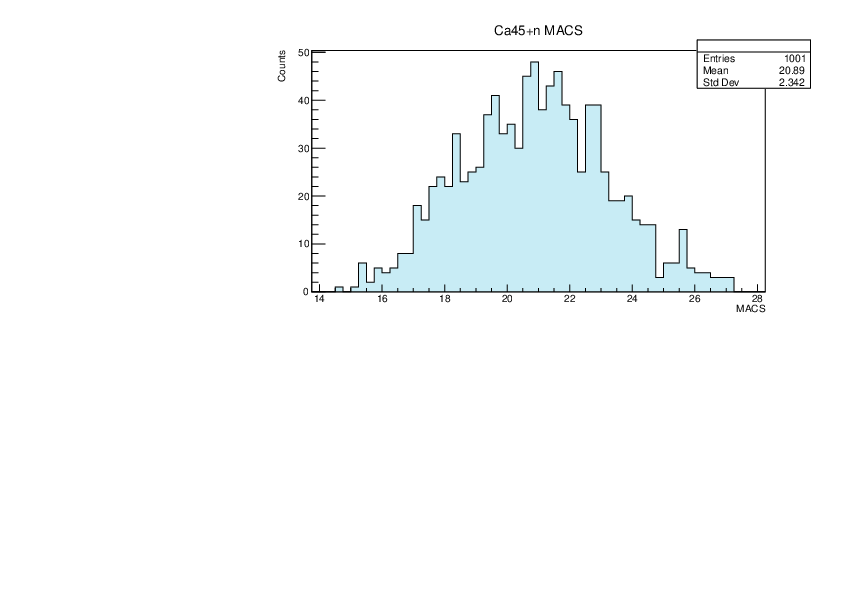
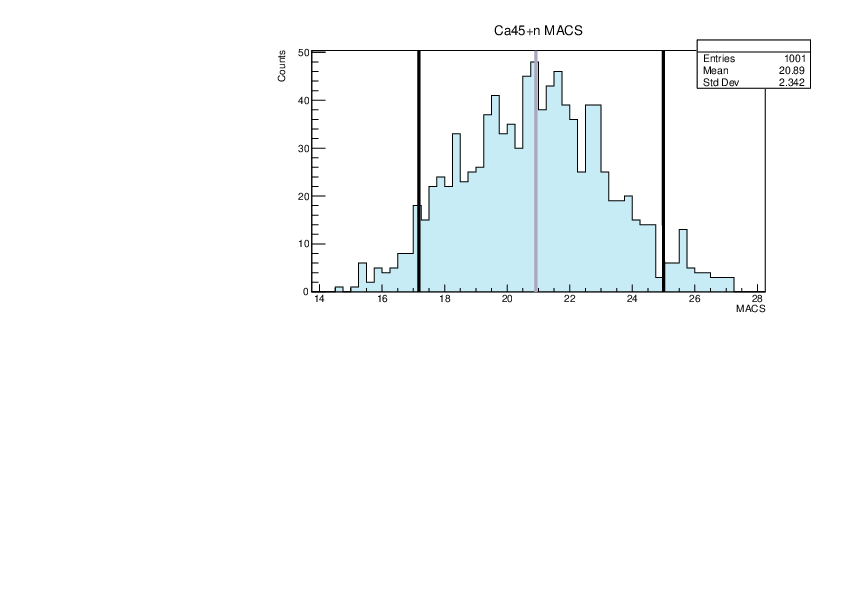
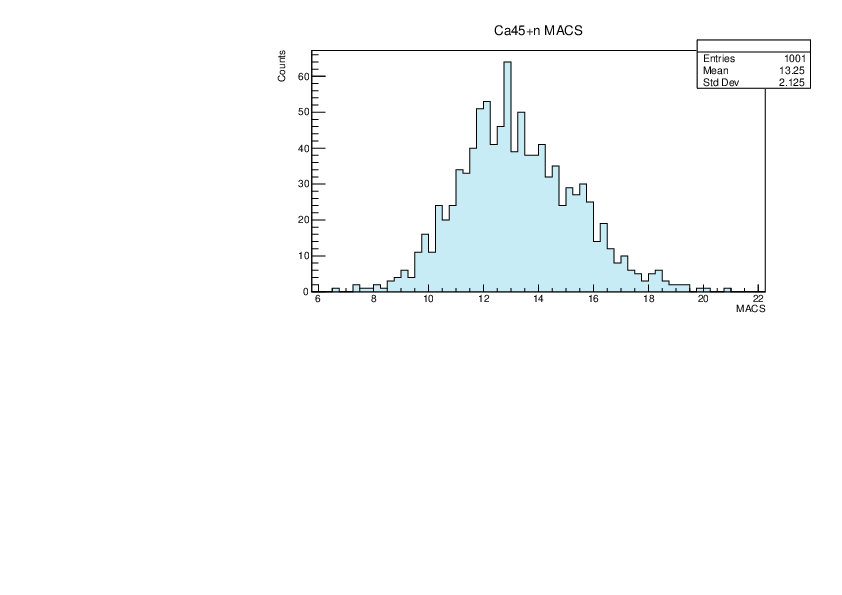
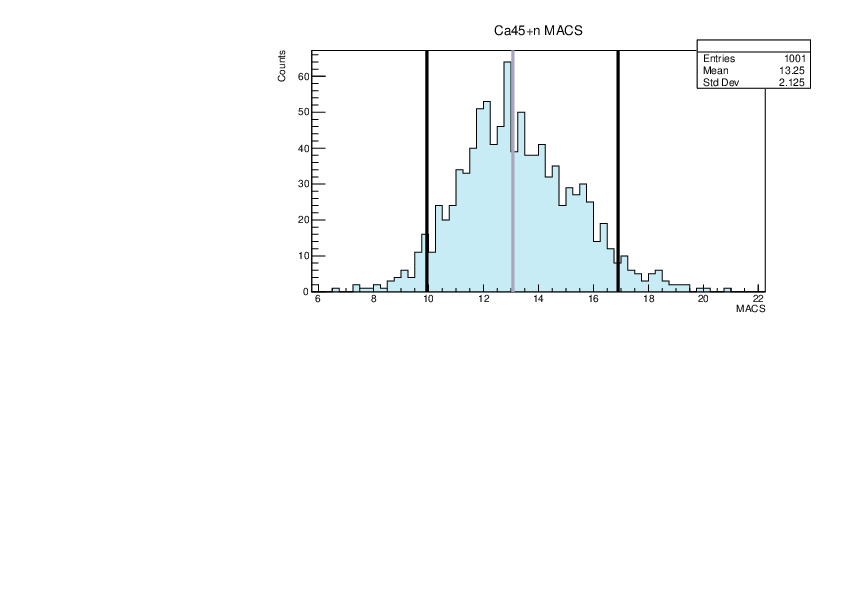
Selin Ezgi Birincioglu | Ca45+n Variations By Combination of Models |
Ca45-20 and -42. |
| Attachment 1: dist.pdf
|
|
| Attachment 2: lim.pdf
|
|
| Attachment 3: dist.pdf
|
|
| Attachment 4: lim.pdf
|
|
|
24
|
Thu Feb 1 10:10:04 2024 |
Selin Ezgi Birincioglu | List of Nuclei with MACS |
This is the Excel sheet with the list of nuclei and their corresponding MACS. Yellow fill shows the nuclei with experimental cross sections, green fill shows the nuclei that obey the statistical calculations and blue fill shows the nuclei with the Gaussian distributions. |
| Attachment 1: rates_clw.xlsx
|
|
22
|
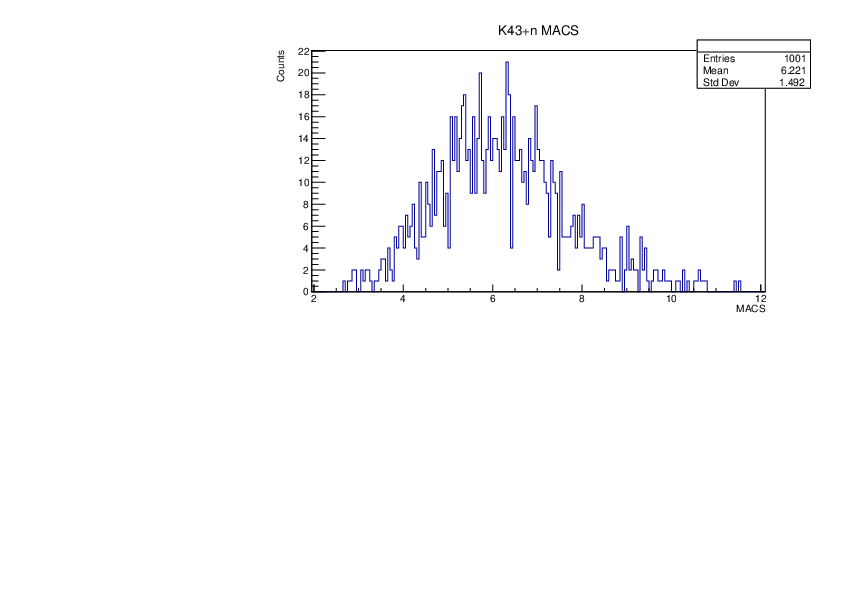
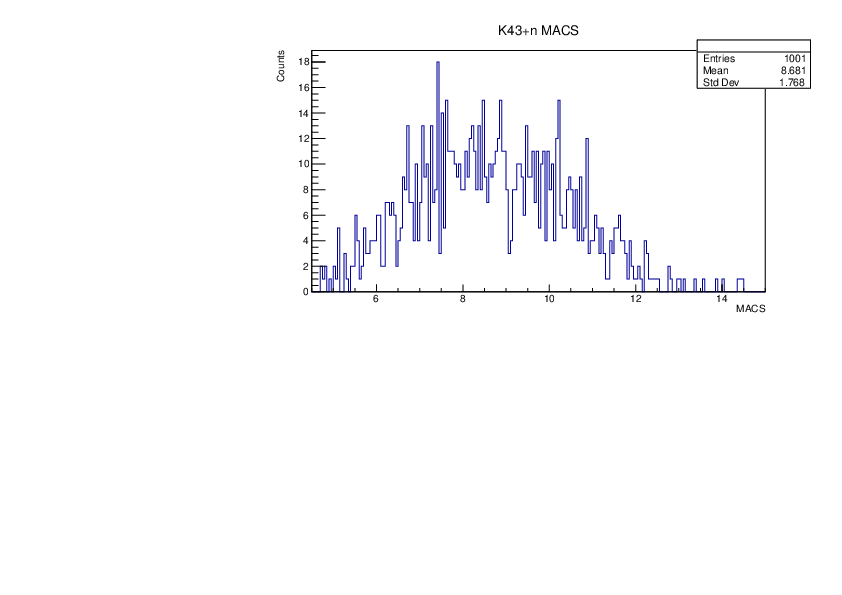
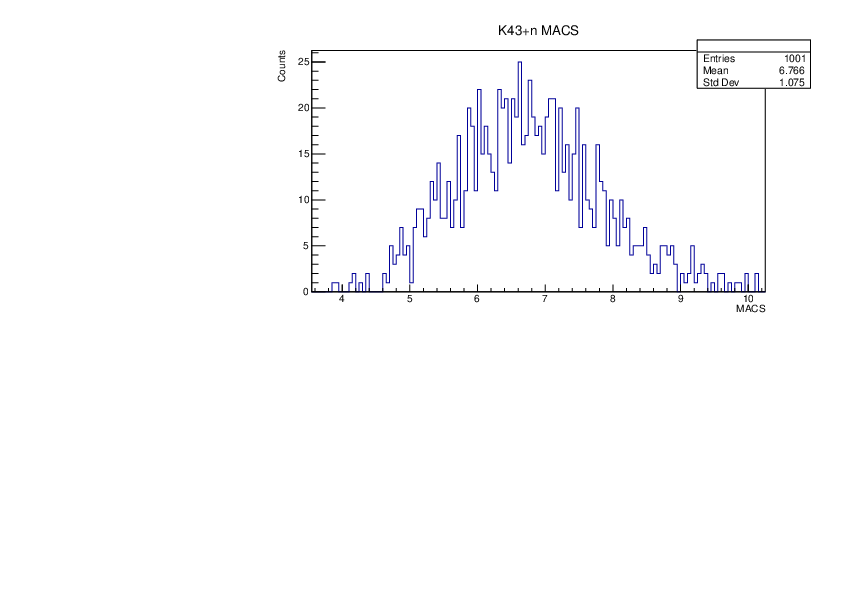
Tue Jan 30 10:37:56 2024 |
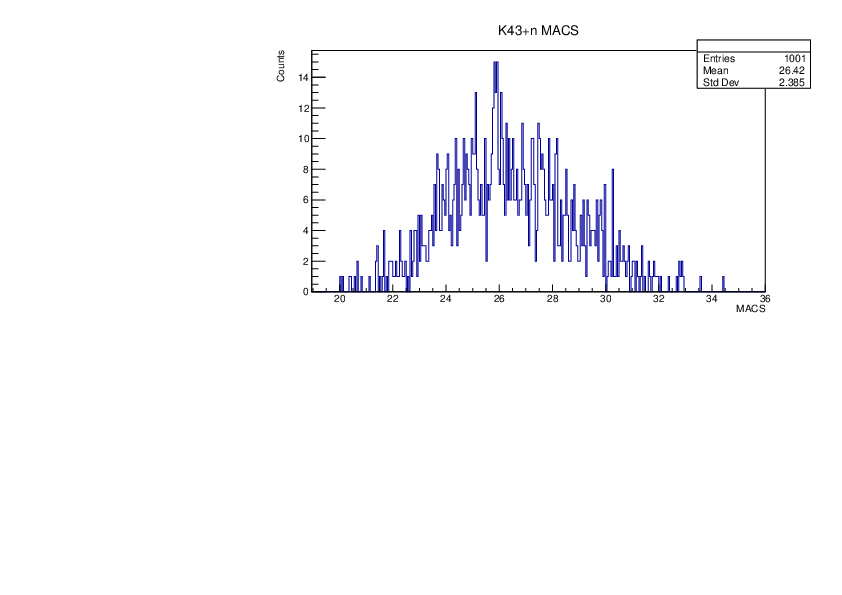
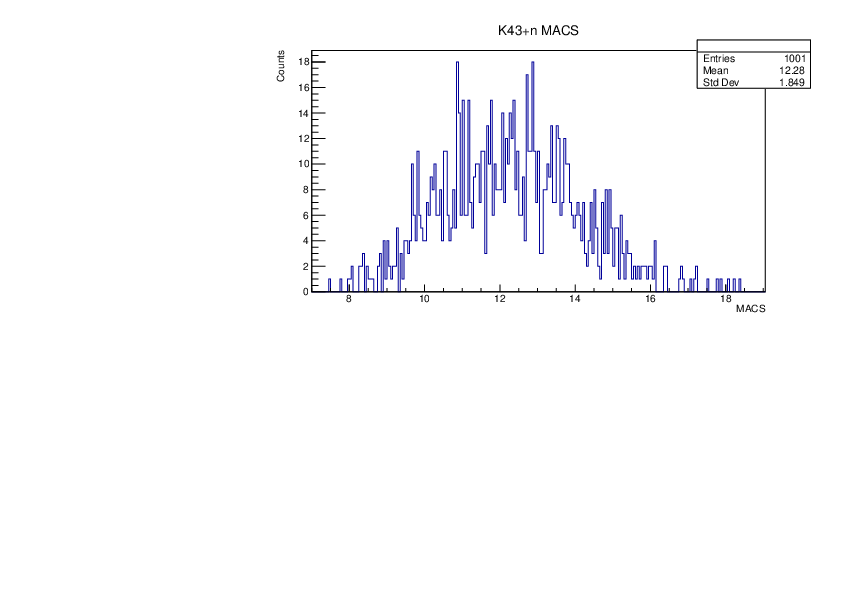
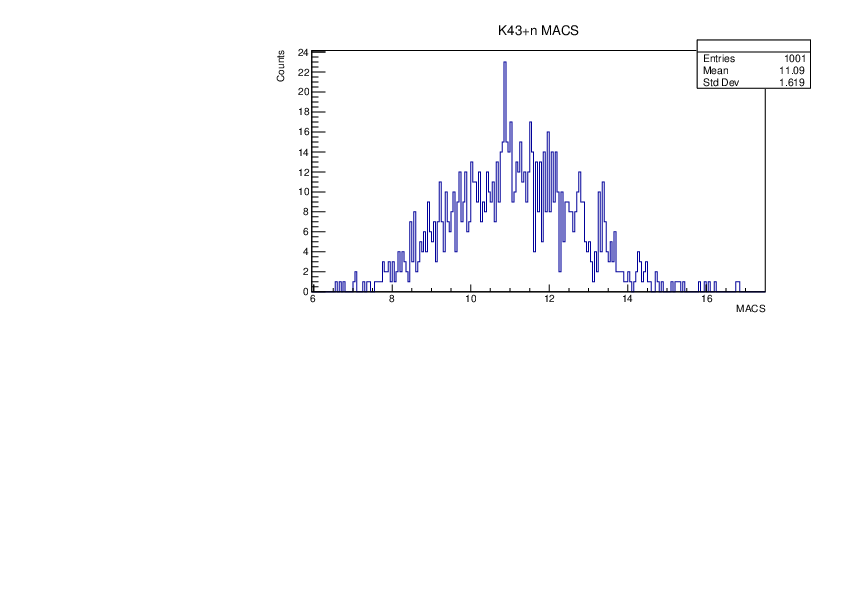
Selin Ezgi Birincioglu | K43+n Variations By Different Models |
The variations of MACS distribution of K43+nwith different level density models. K43+n cannot be predicted by the statistical model below 0.025 MeV. |
| Attachment 1: ld1.pdf
|
|
| Attachment 2: ld2.pdf
|
|
| Attachment 3: ld3.pdf
|
|
| Attachment 4: ld4.pdf
|
|
| Attachment 5: ld5.pdf
|
|
| Attachment 6: ld6.pdf
|
|
|
21
|
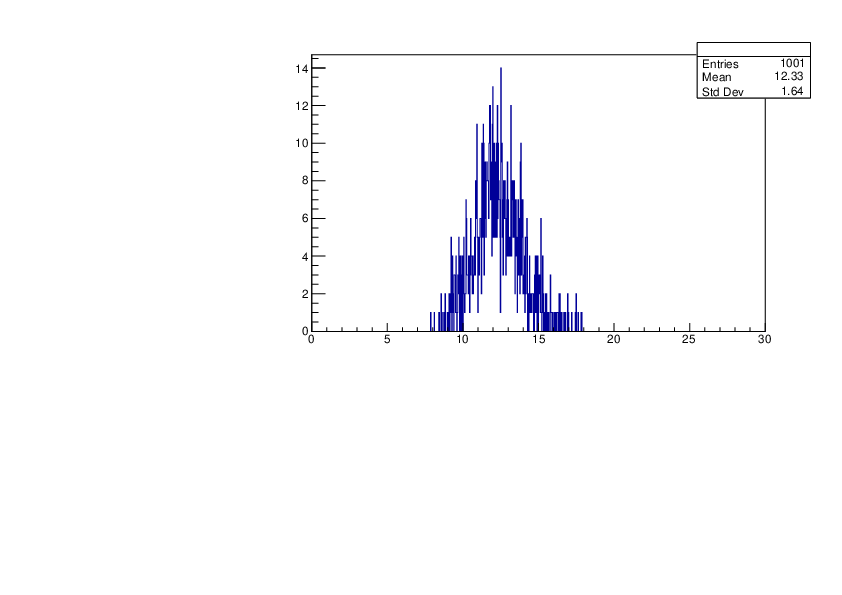
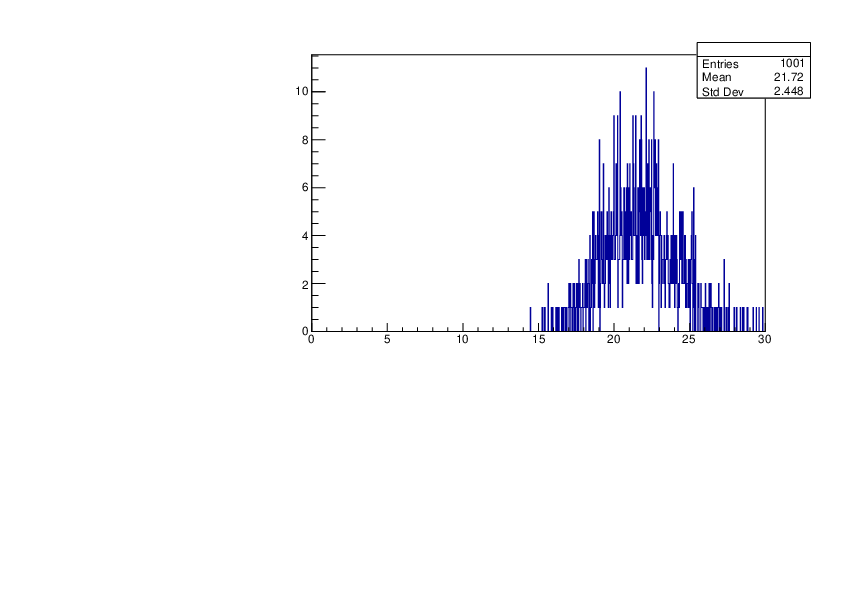
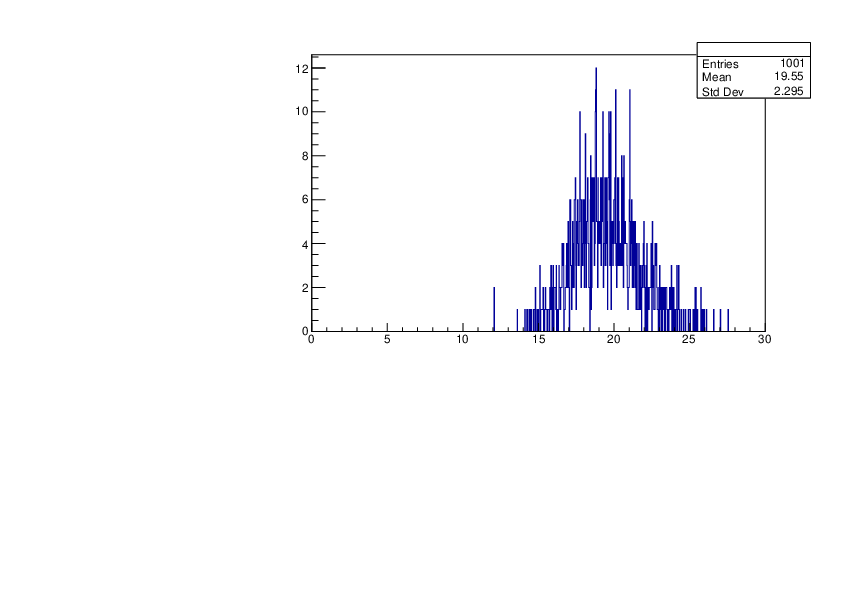
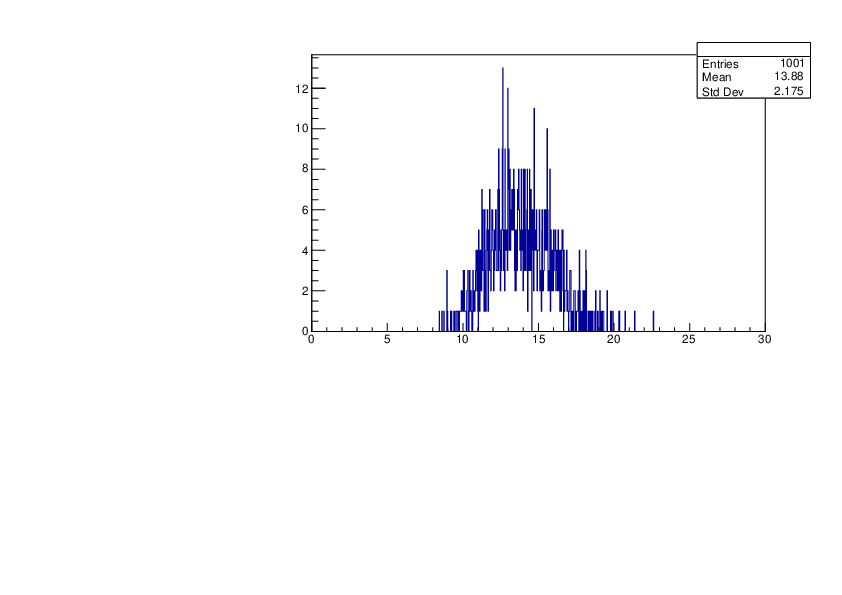
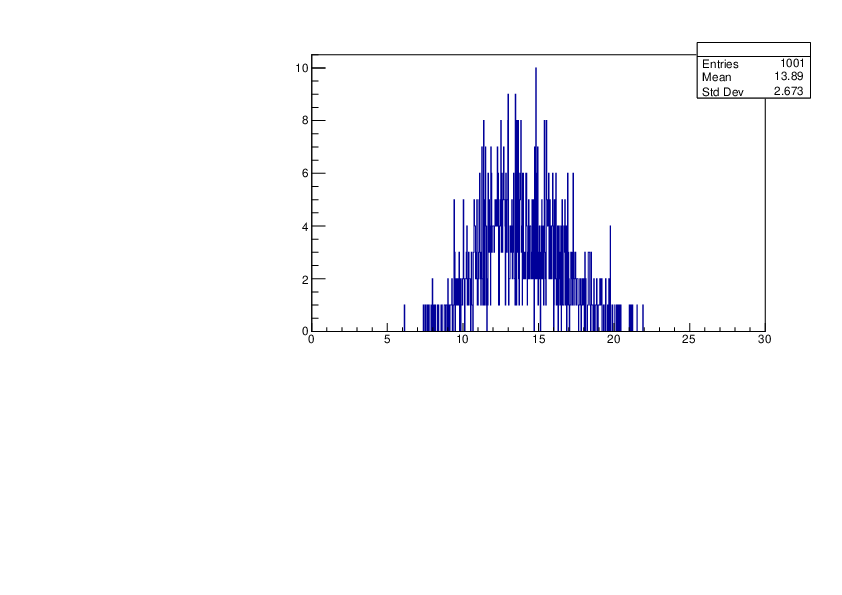
Thu Jan 18 17:36:32 2024 |
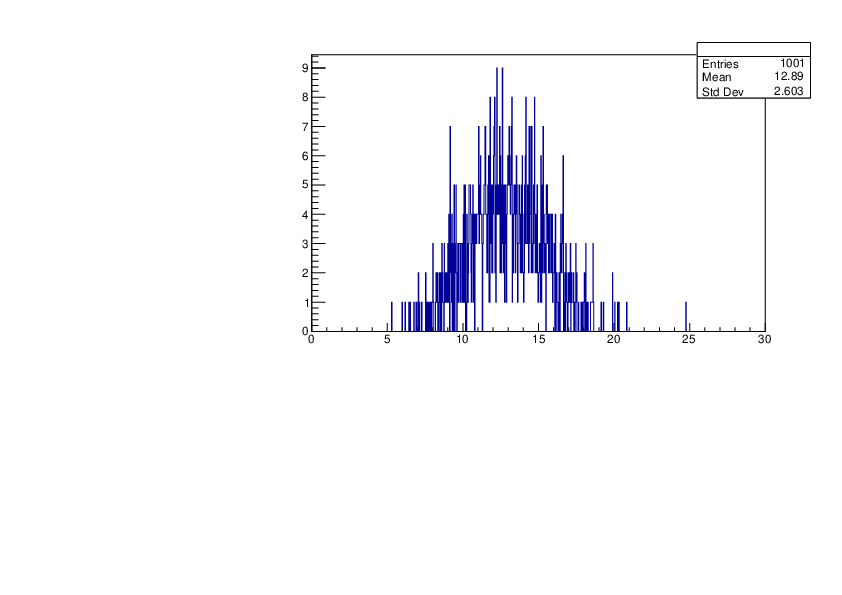
Selin Ezgi Birincioglu | Ca45+n Variations By Different Models |
The variations of MACS distribution of Ca45+n with different level density models. Ca45+n cannot be predicted by the statistical model below approx. 0.020 MeV. |
| Attachment 1: ld1.pdf
|
|
| Attachment 2: ld2.pdf
|
|
| Attachment 3: ld3.pdf
|
|
| Attachment 4: ld4.pdf
|
|
| Attachment 5: ld5.pdf
|
|
| Attachment 6: ld6.pdf
|
|
|
20
|
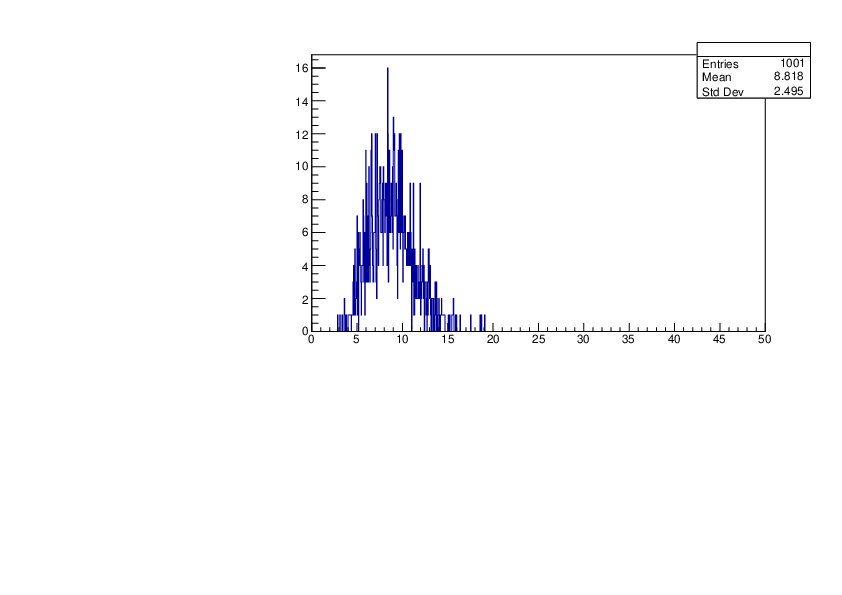
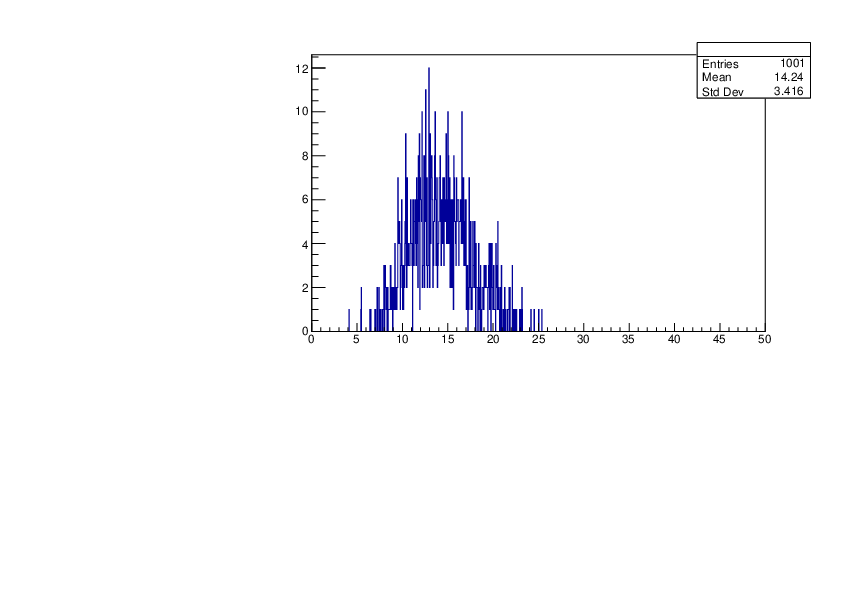
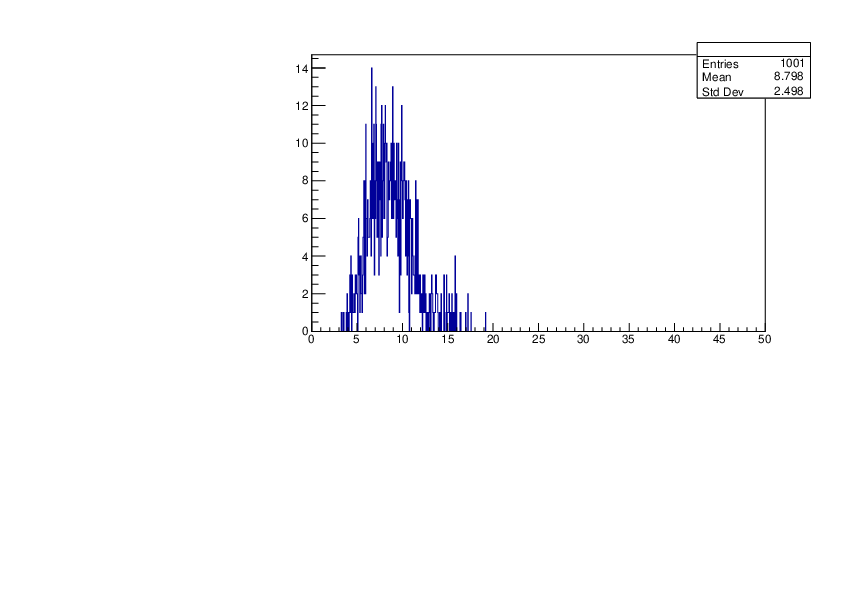
Thu Jan 18 15:43:13 2024 |
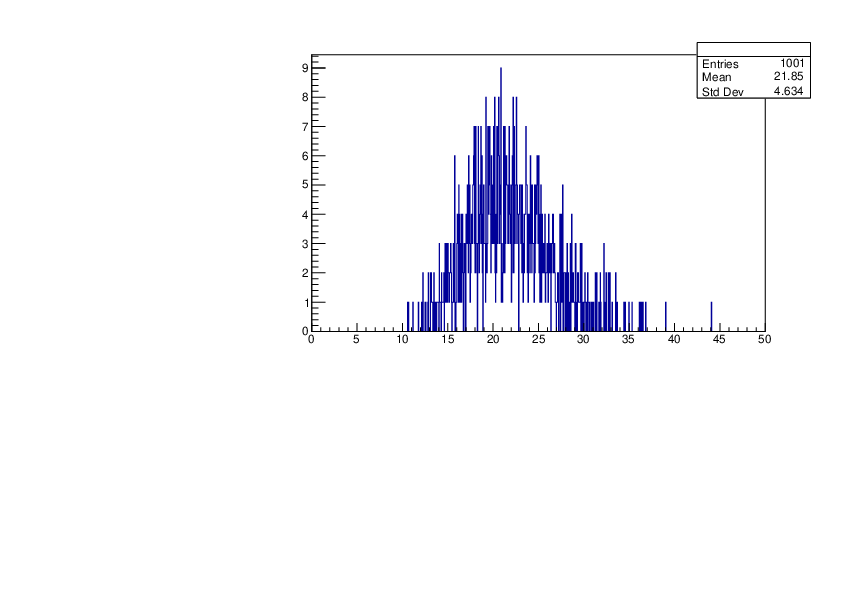
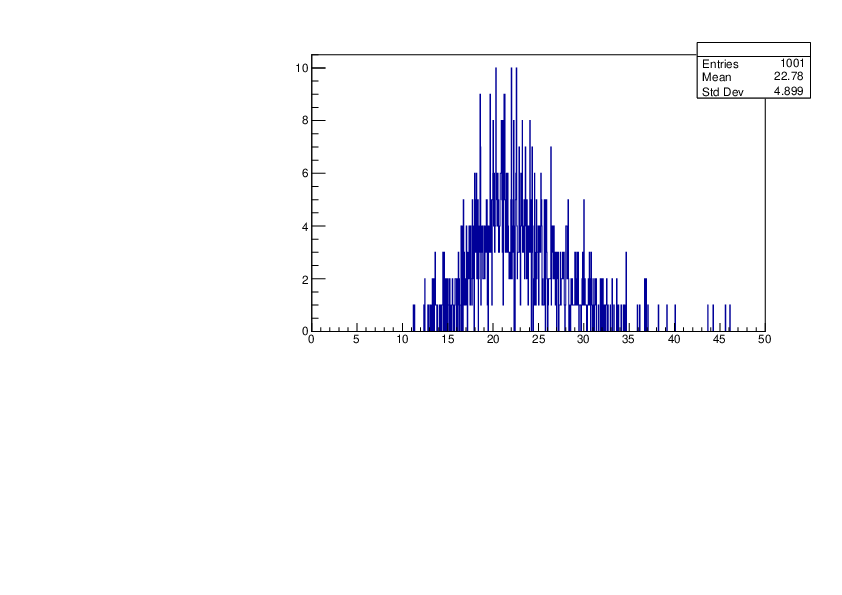
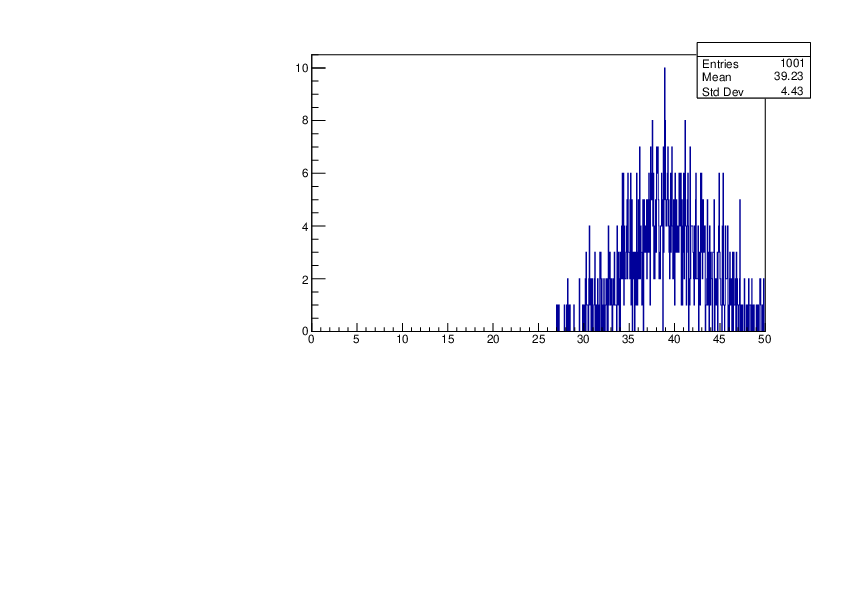
Selin Ezgi Birincioglu | K41+n Variations By Different Models |
The variations of MACS distribution of K41+nwith different level density models. K41+n cannot be predicted by the statistical model below 0.022 MeV. |
| Attachment 1: ld1.pdf
|
|
| Attachment 2: ld2.pdf
|
|
| Attachment 3: ld3.pdf
|
|
| Attachment 4: ld4.pdf
|
|
| Attachment 5: ld5.pdf
|
|
| Attachment 6: ld6.pdf
|
|
|
19
|
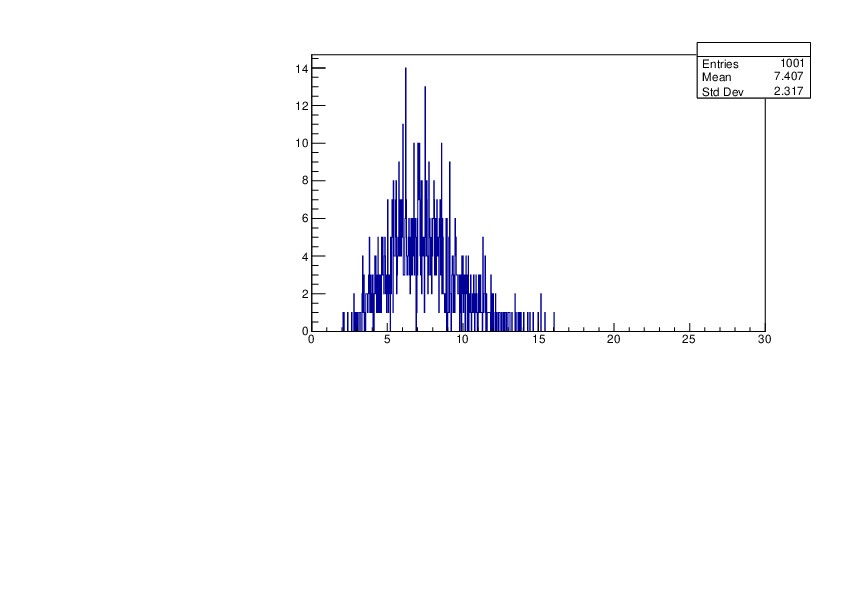
Thu Jan 18 11:08:23 2024 |
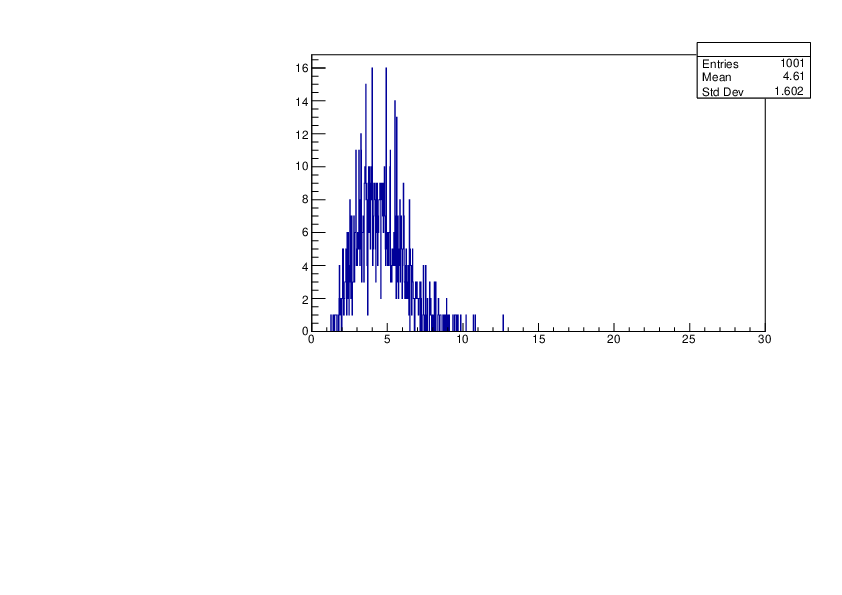
Selin Ezgi Birincioglu | K39+n Variations By Different Models |
The variations of MACS distribution of K39+nwith different level density models. K39+ncannot be predicted by the statistical model below 0.028 MeV. |
| Attachment 1: ld1.pdf
|
|
| Attachment 2: ld2.pdf
|
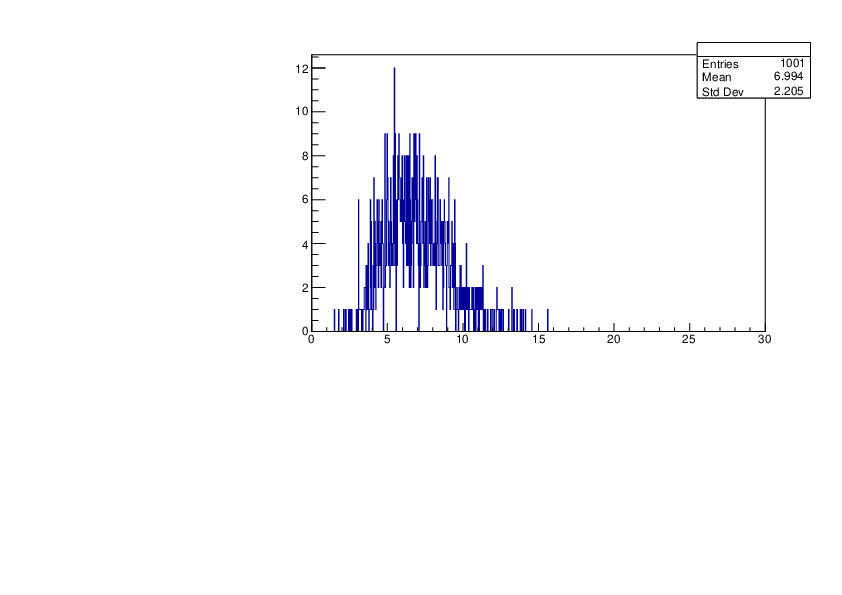
|
| Attachment 3: ld3.pdf
|
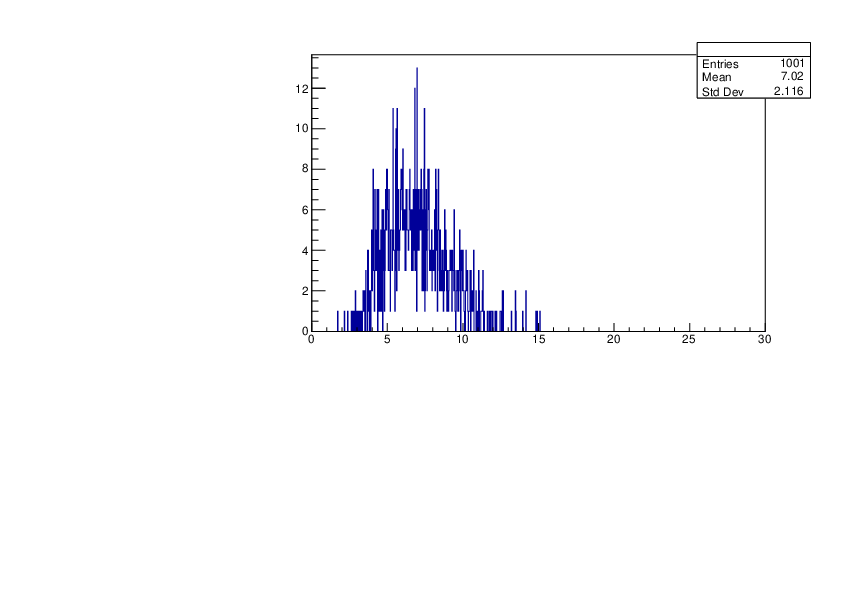
|
| Attachment 4: ld4.pdf
|
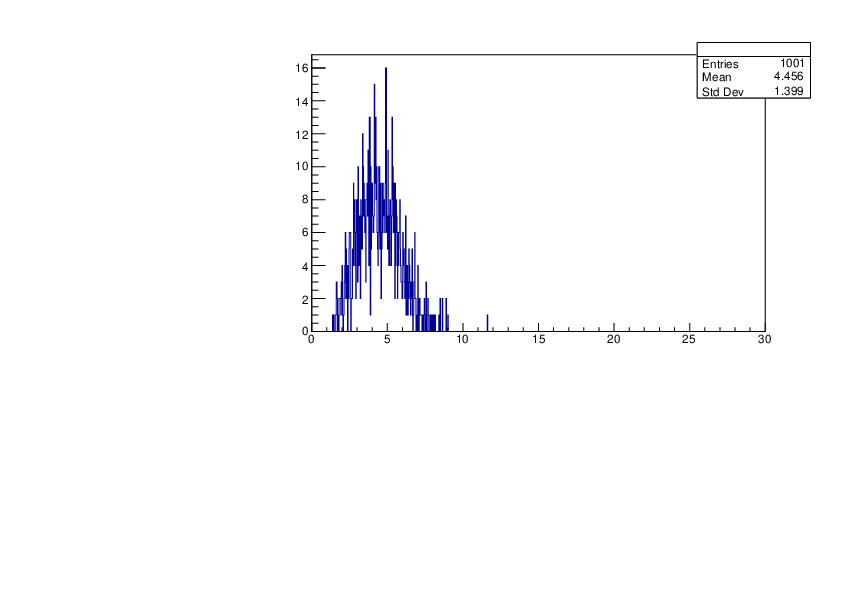
|
| Attachment 5: ld5.pdf
|
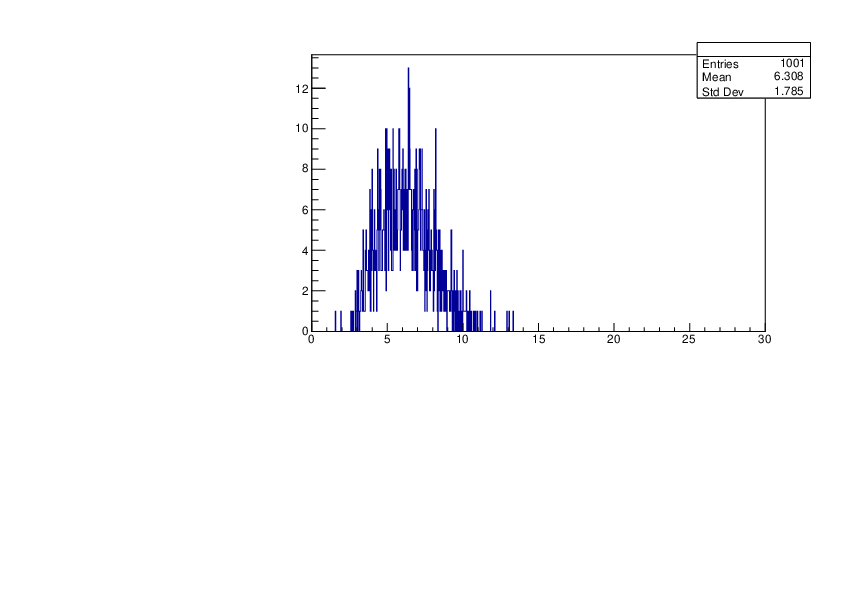
|
| Attachment 6: ld6.pdf
|
|
|
18
|
Thu Jan 18 10:52:48 2024 |
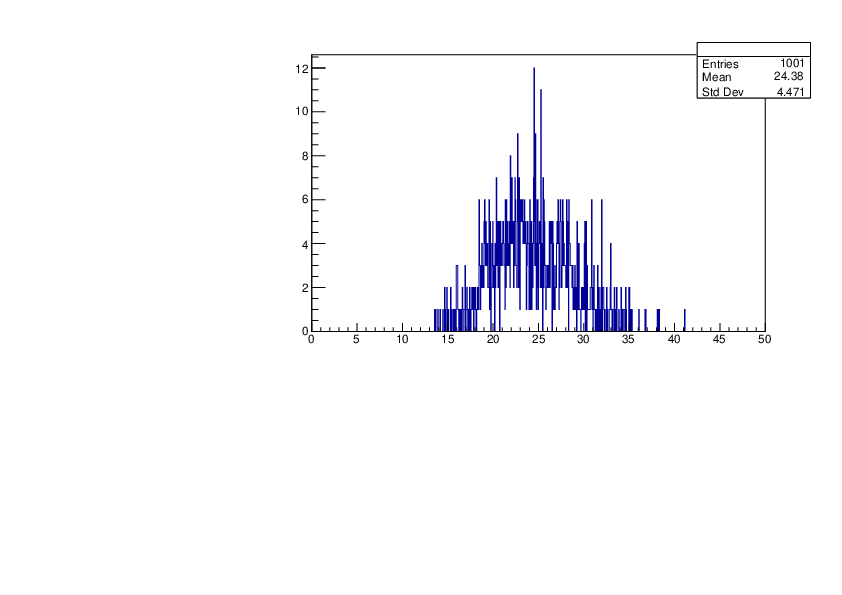
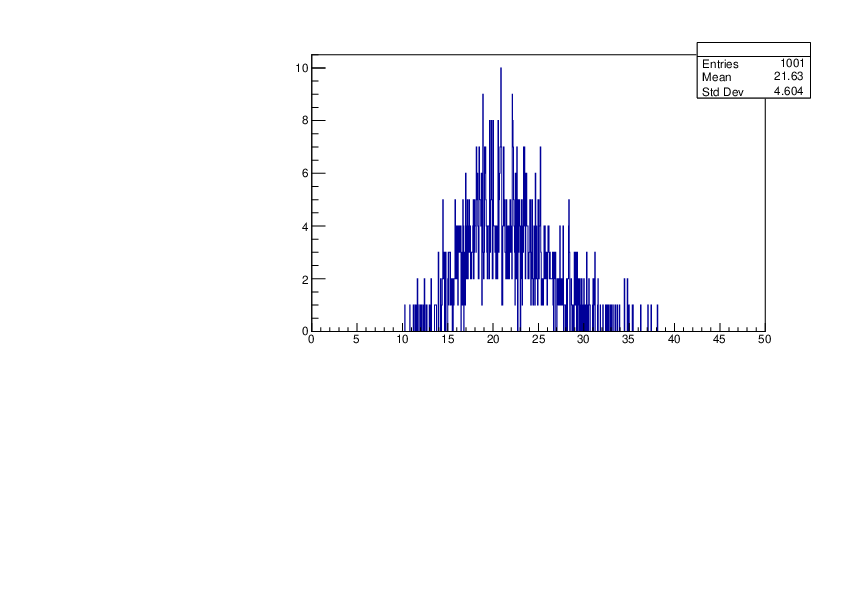
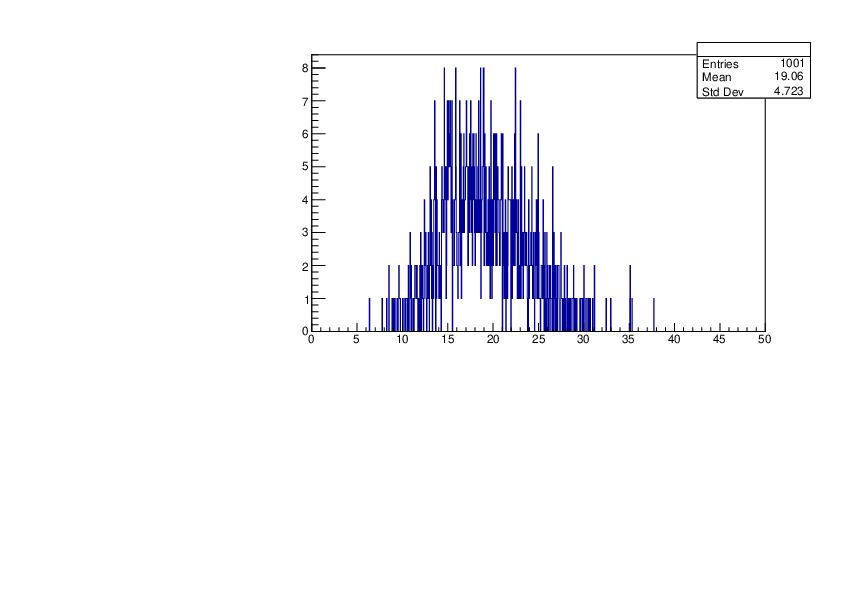
Selin Ezgi Birincioglu | Ar37+n Variations by Different Models |
The variations of MACS distribution of Ar37+n with different level density models. Ar37+n cannot be predicted by the statistical model below 0.022 MeV. |
| Attachment 1: ld1.pdf
|
|
| Attachment 2: ld2.pdf
|
|
| Attachment 3: ld3.pdf
|
|
| Attachment 4: ld4.pdf
|
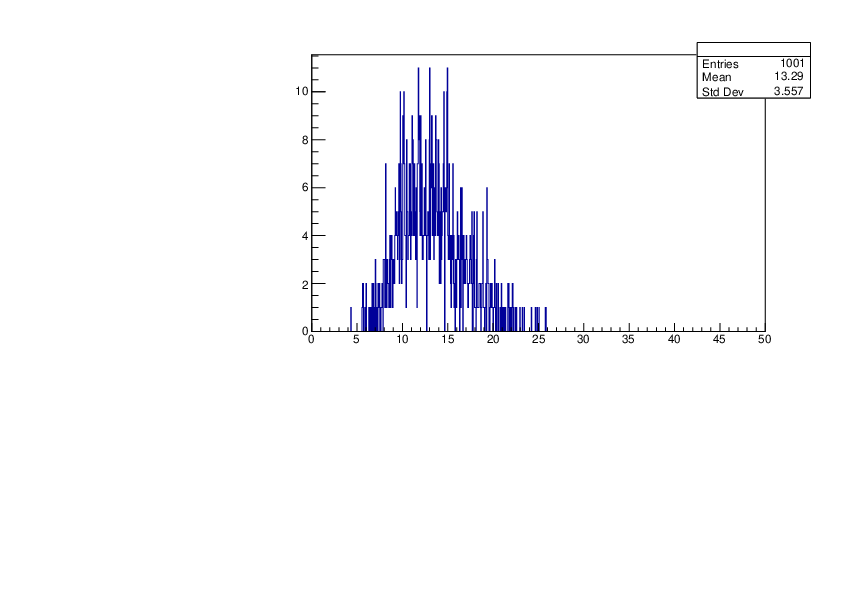
|
| Attachment 5: ld5.pdf
|
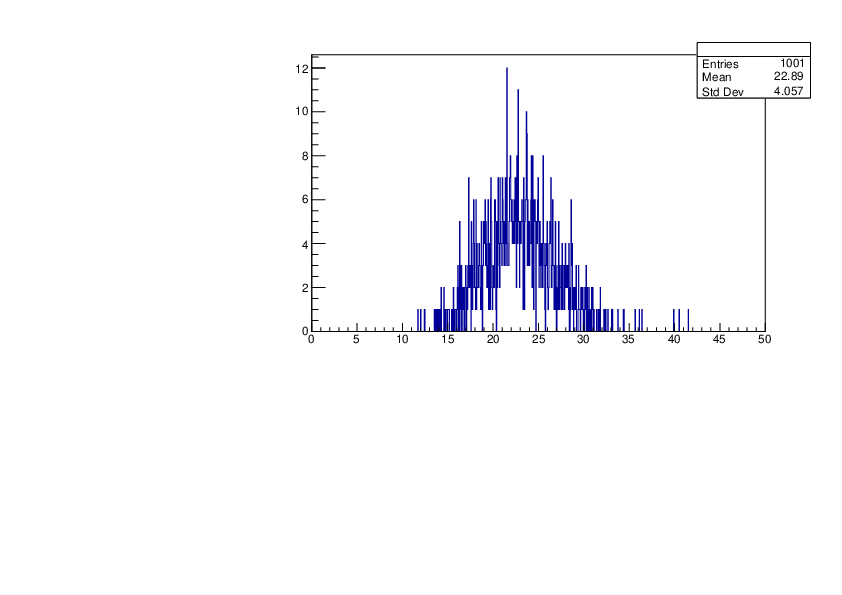
|
| Attachment 6: ld6.pdf
|
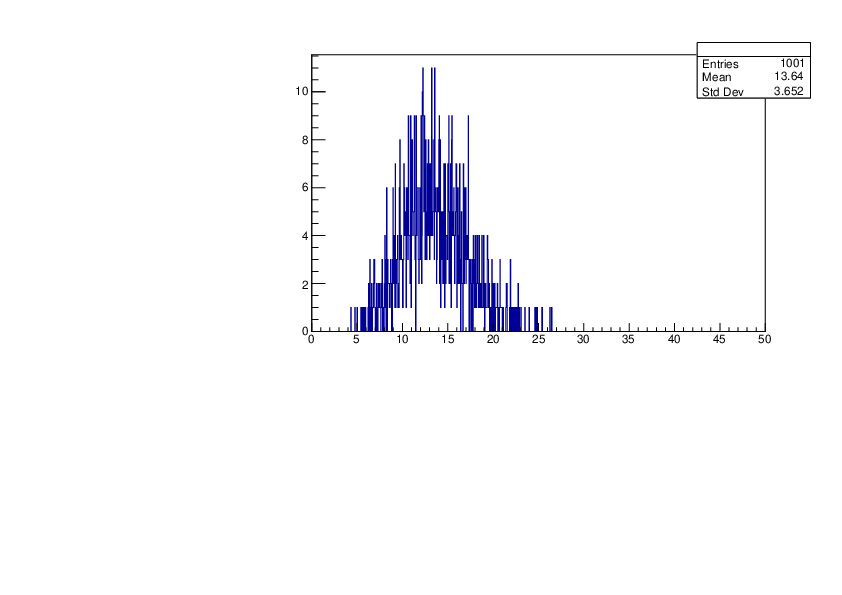
|
|
17
|
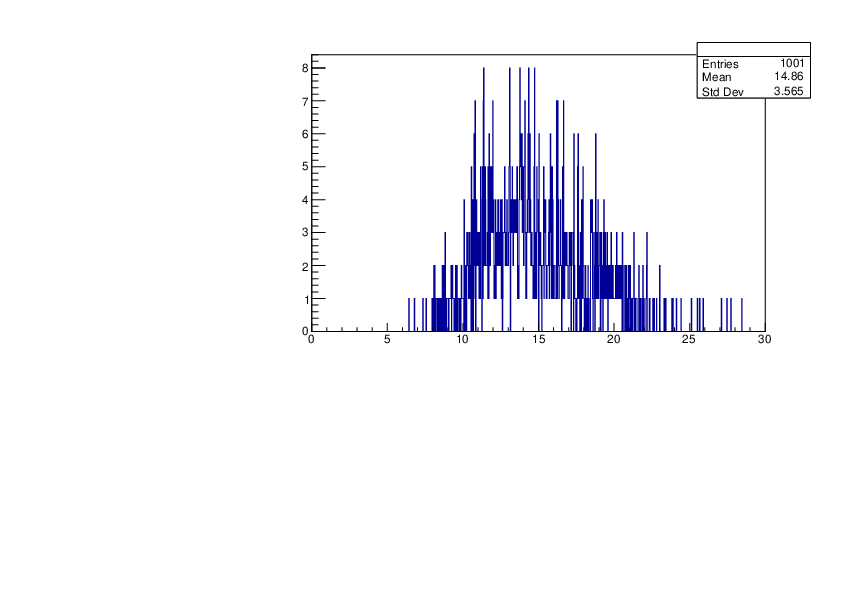
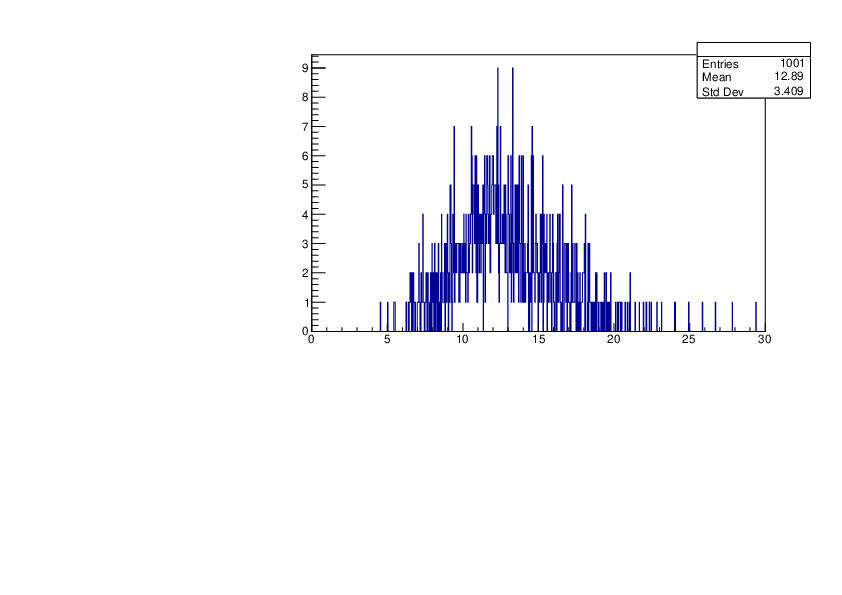
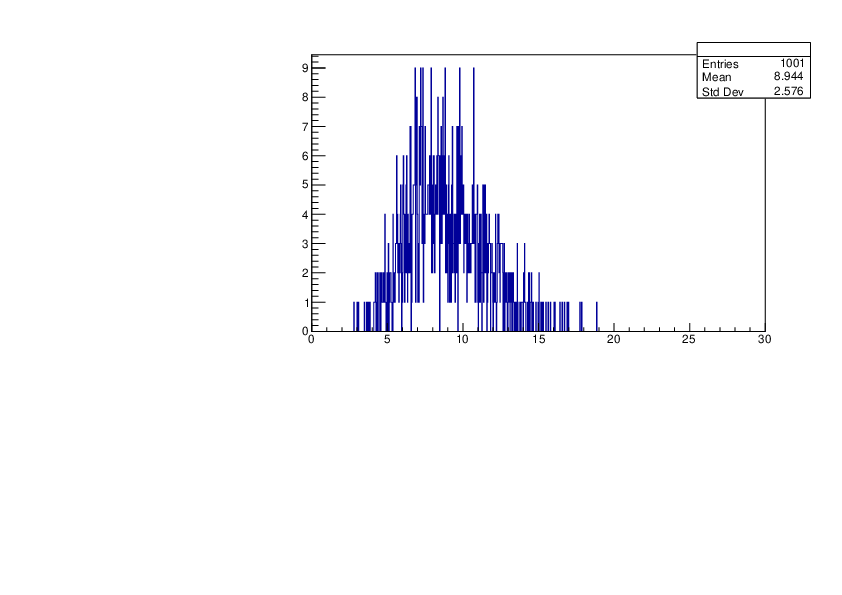
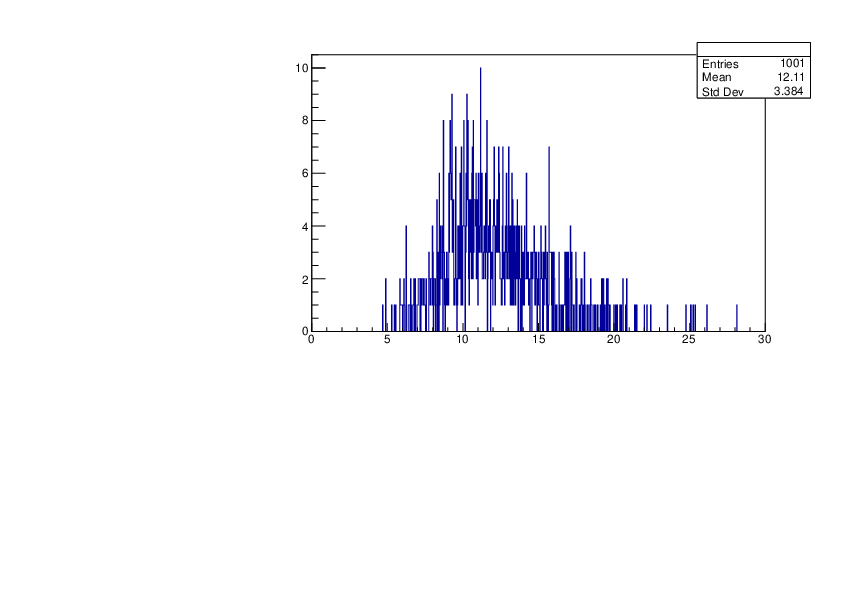
Wed Jan 17 16:15:49 2024 |
Selin Ezgi Birincioglu | Cl36+n Variations by Different Models |
The variations of MACS distribution of Cl36+n with different level density models. Cl36+n cannot be predicted by the statistical model below approx. 0.020 MeV. |
| Attachment 1: ld1.pdf
|
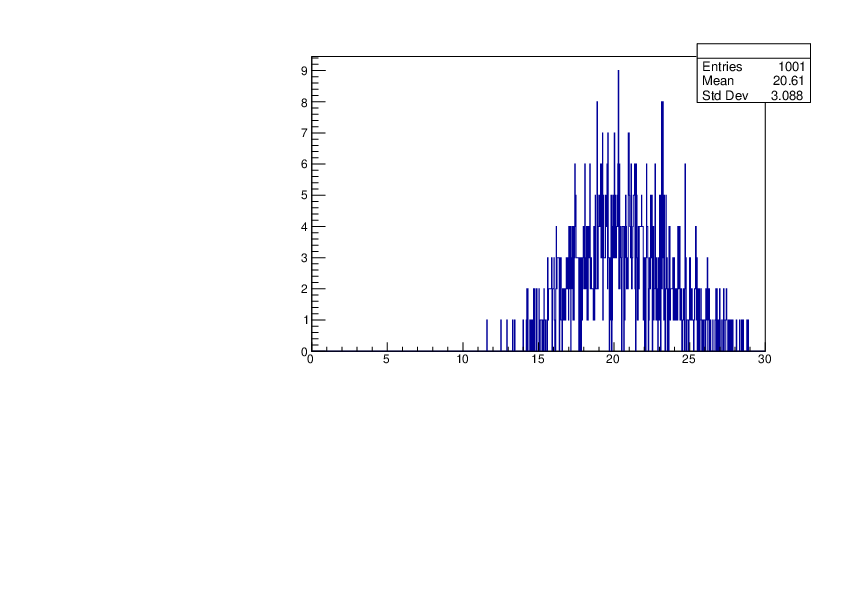
|
| Attachment 2: ld2.pdf
|
|
| Attachment 3: ld3.pdf
|
|
| Attachment 4: ld4.pdf
|
|
| Attachment 5: ld5.pdf
|
|
| Attachment 6: ld6.pdf
|
|
|
16
|
Wed Jan 17 15:58:40 2024 |
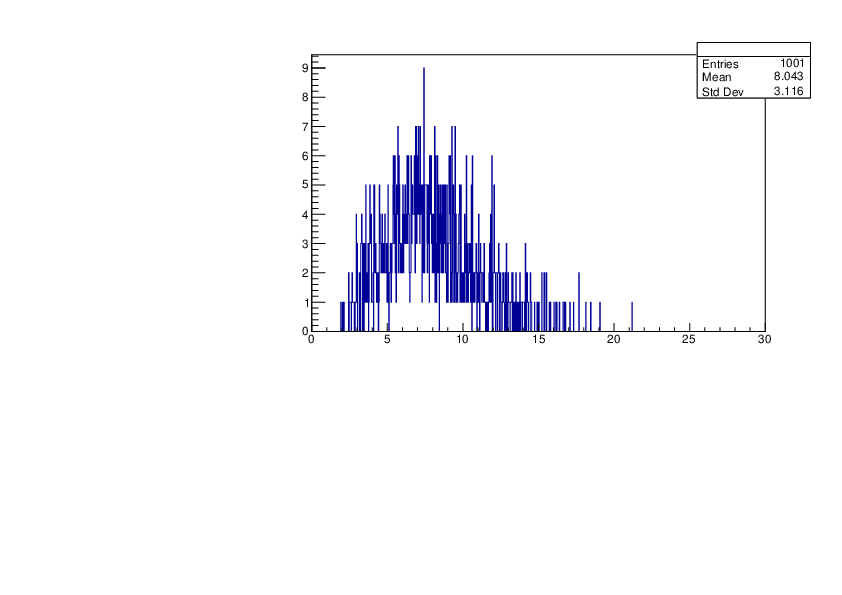
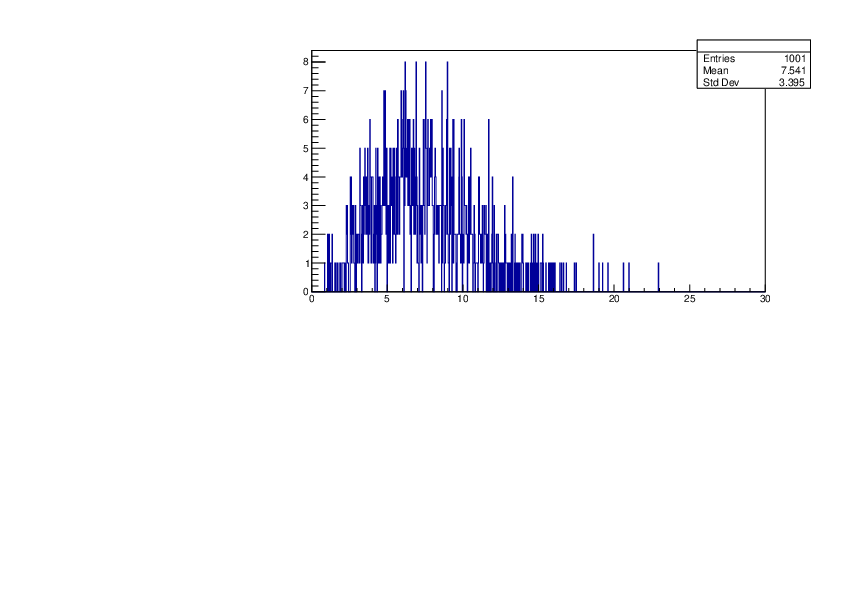
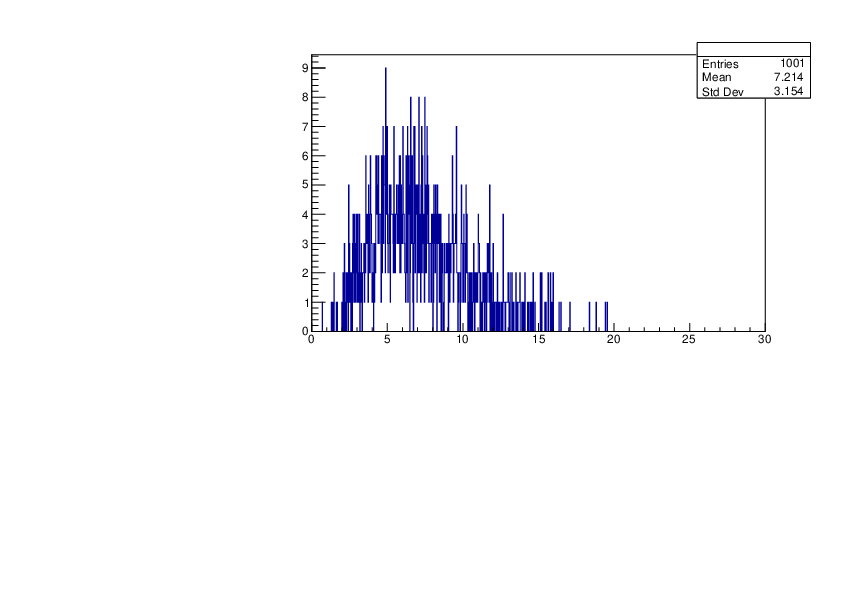
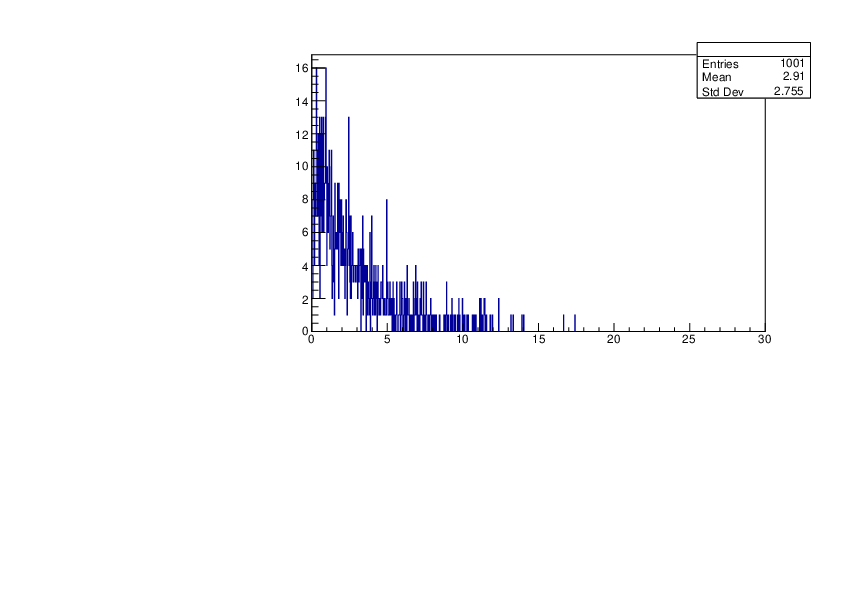
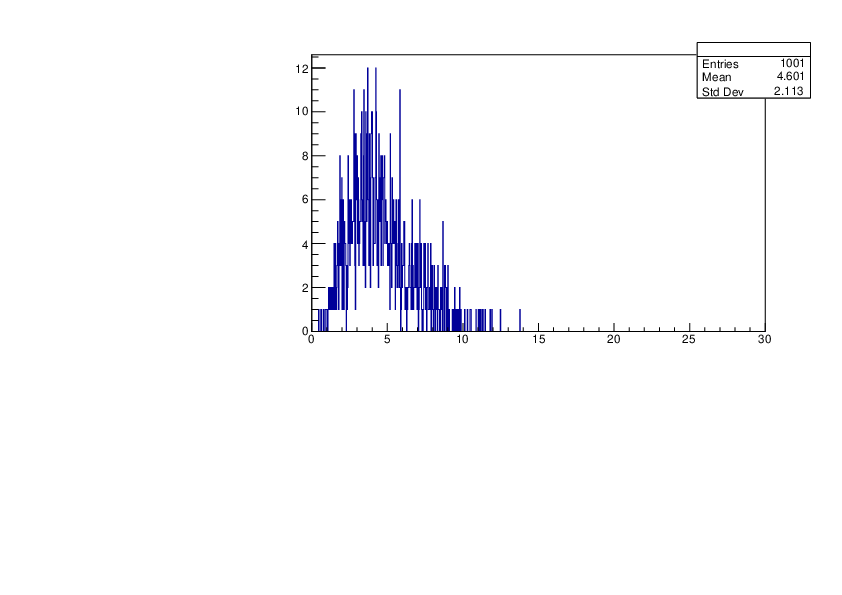
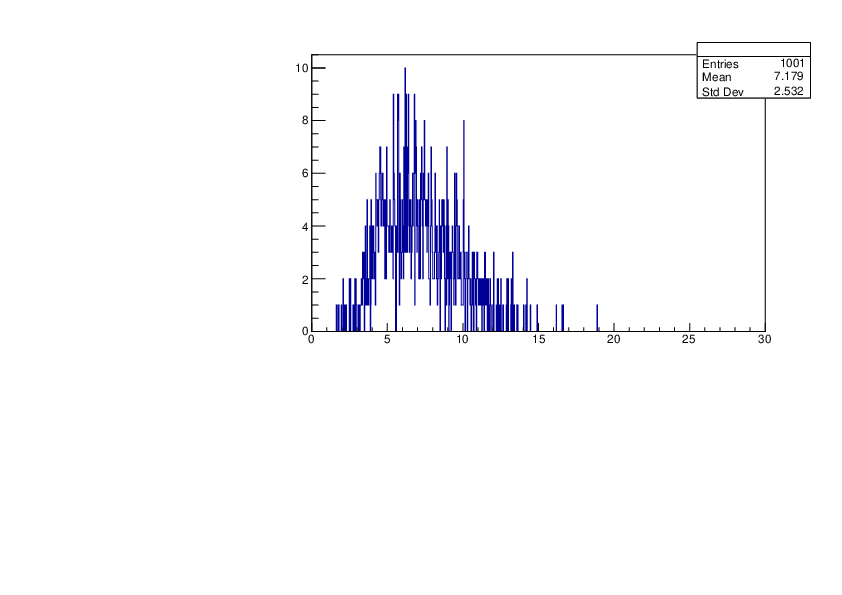
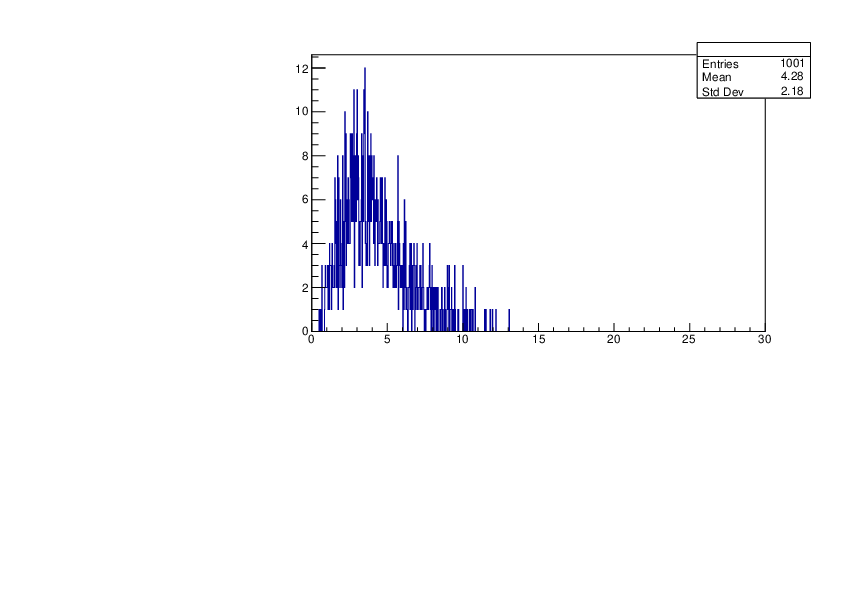
Selin Ezgi Birincioglu | Cl35+n Variations by Different Models |
The variations of MACS distribution of Cl35+n with different level density models. Cl35+n cannot be predicted by the statistical model belowe 0.026 MeV. |
| Attachment 1: ld1.pdf
|
|
| Attachment 2: ld2.pdf
|
|
| Attachment 3: ld3.pdf
|
|
| Attachment 4: ld4.pdf
|
|
| Attachment 5: ld5.pdf
|
|
| Attachment 6: ld6.pdf
|
|
|
15
|
Wed Jan 17 09:23:28 2024 |
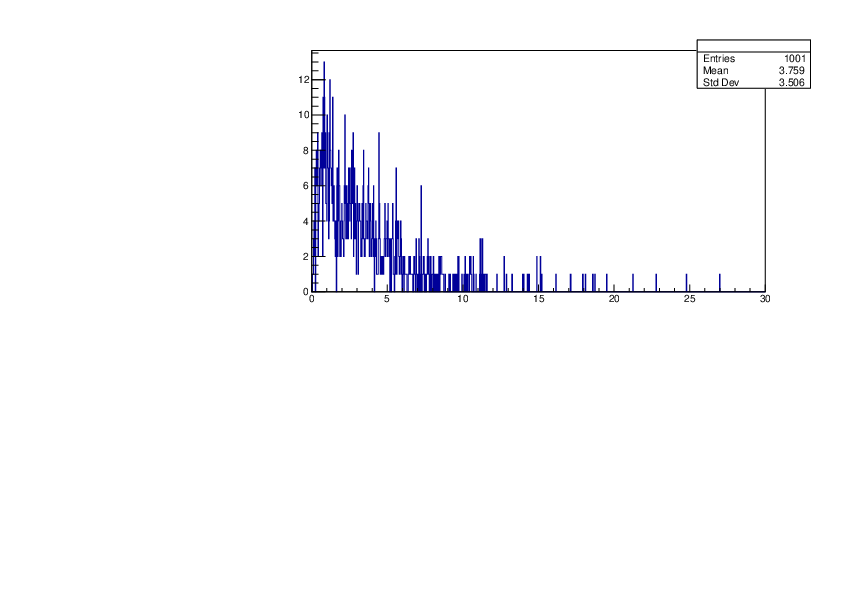
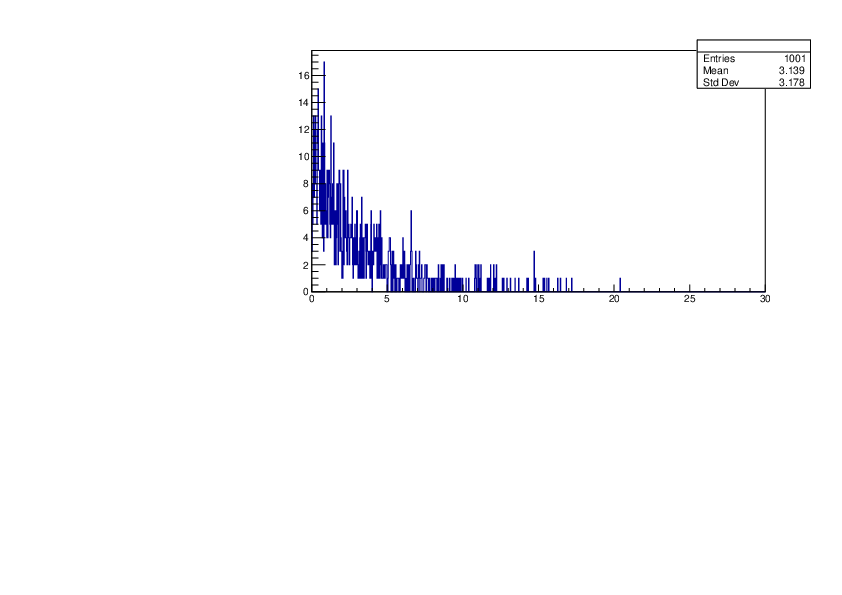
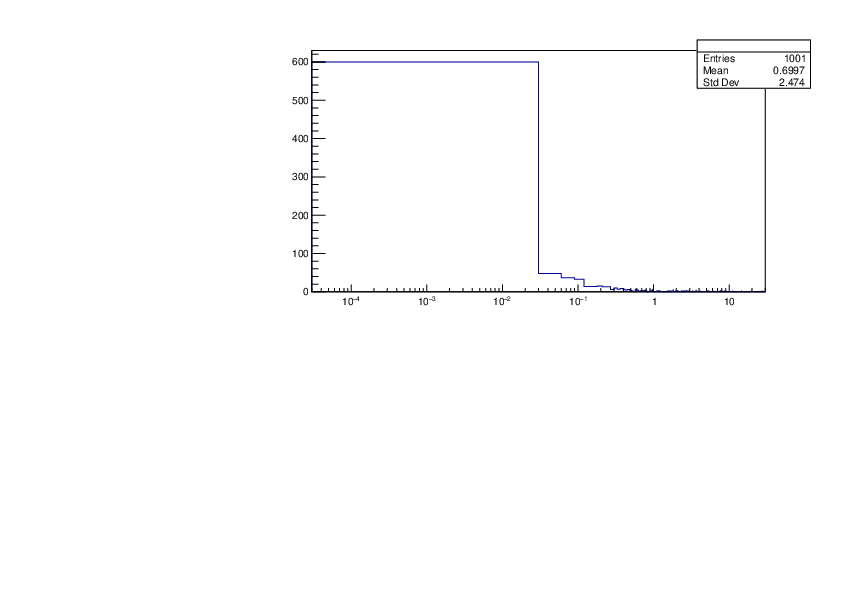
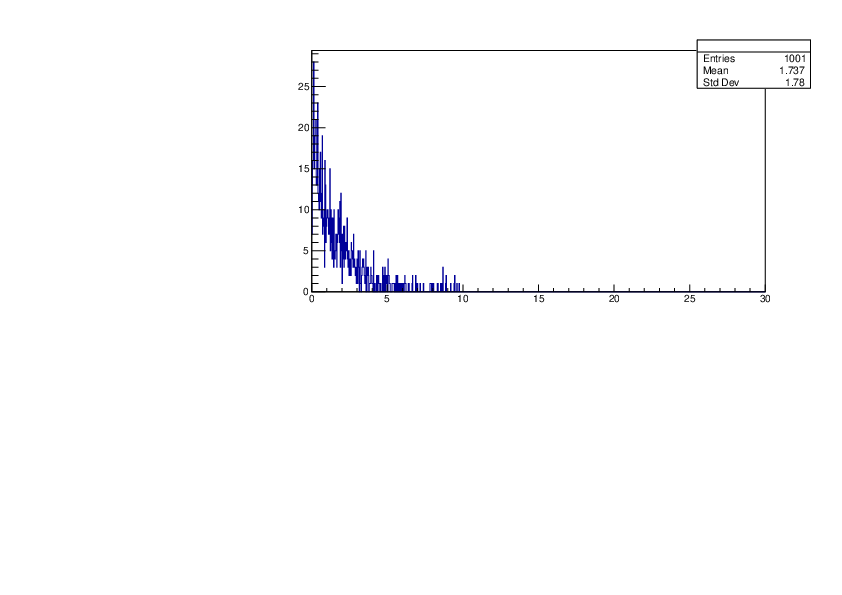
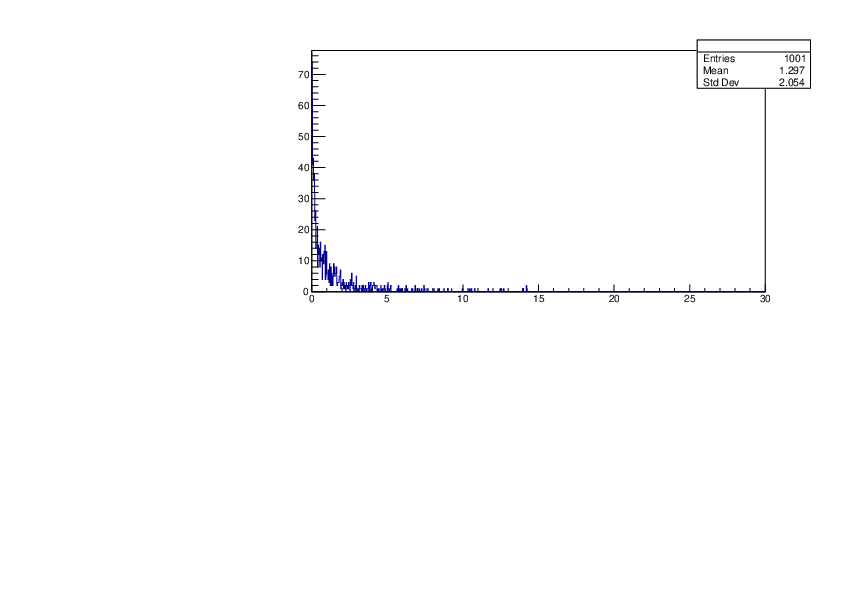
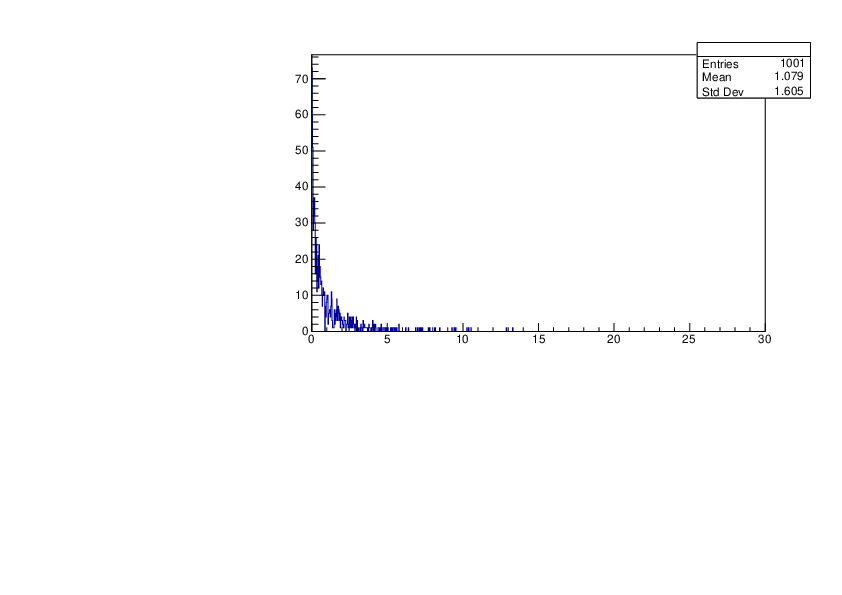
Selin Ezgi Birincioglu | P31+n Variations By Different Models |
The variations of MACS distribution of P31+n with different nuclear models generated by TALYS. |
| Attachment 1: ld1.pdf
|
|
| Attachment 2: ld2.pdf
|
|
| Attachment 3: ld3.pdf
|
|
| Attachment 4: ld4.pdf
|
|
| Attachment 5: ld5.pdf
|
|
| Attachment 6: ld6.pdf
|
|
|
14
|
Wed Jan 17 09:17:57 2024 |
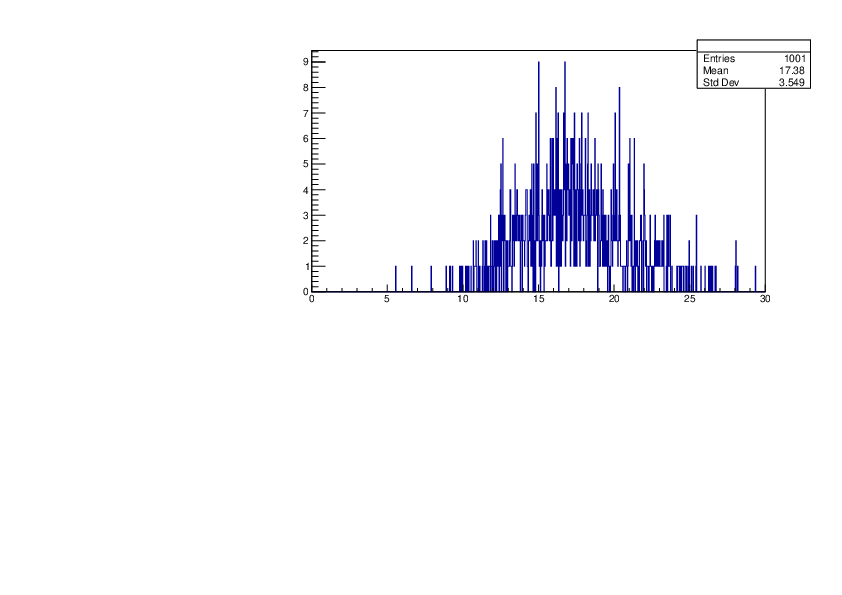
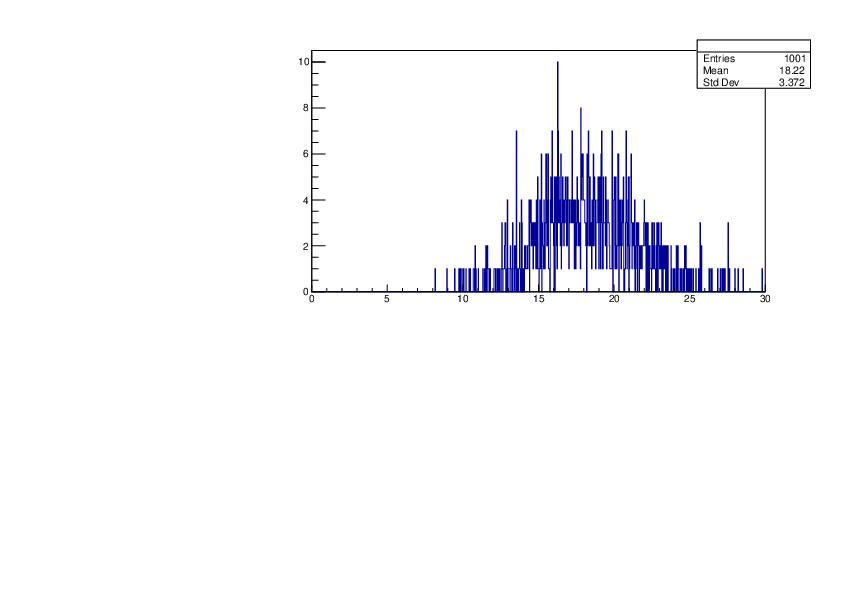
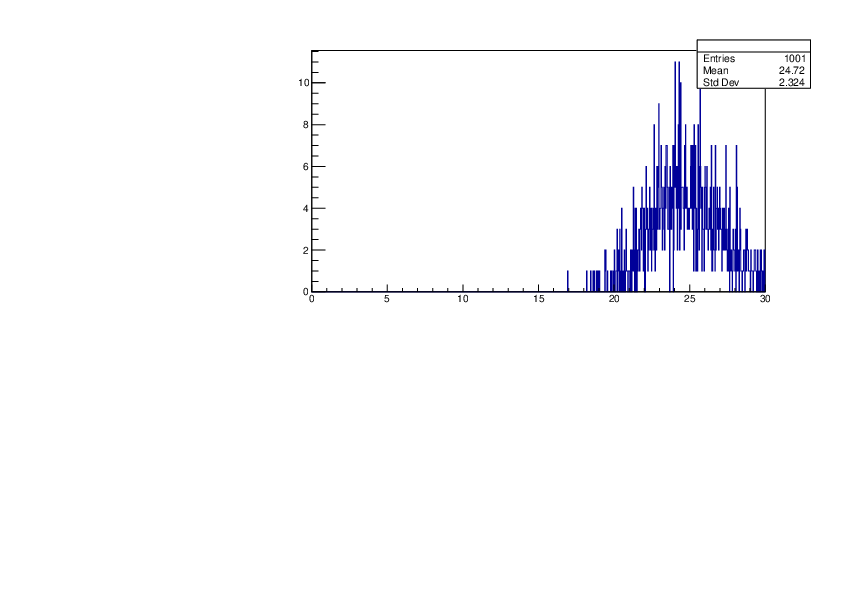
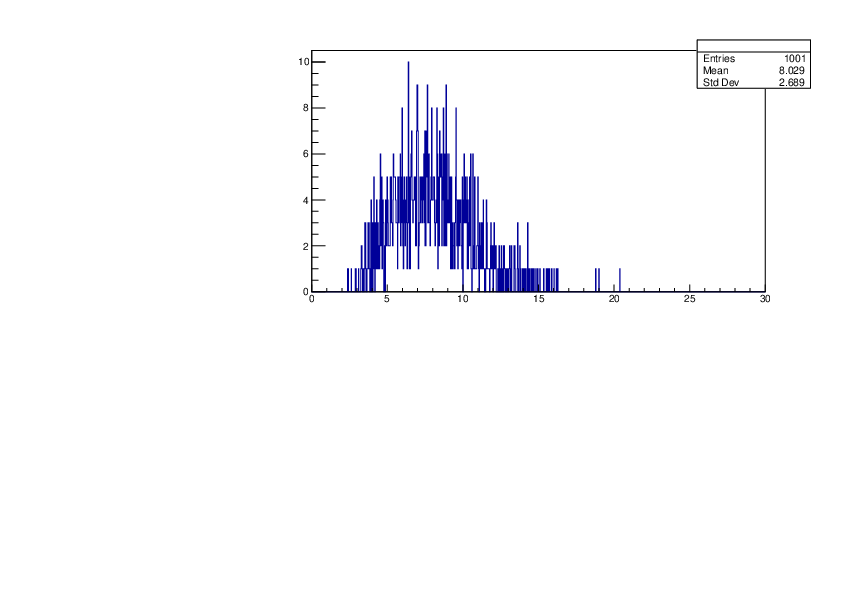
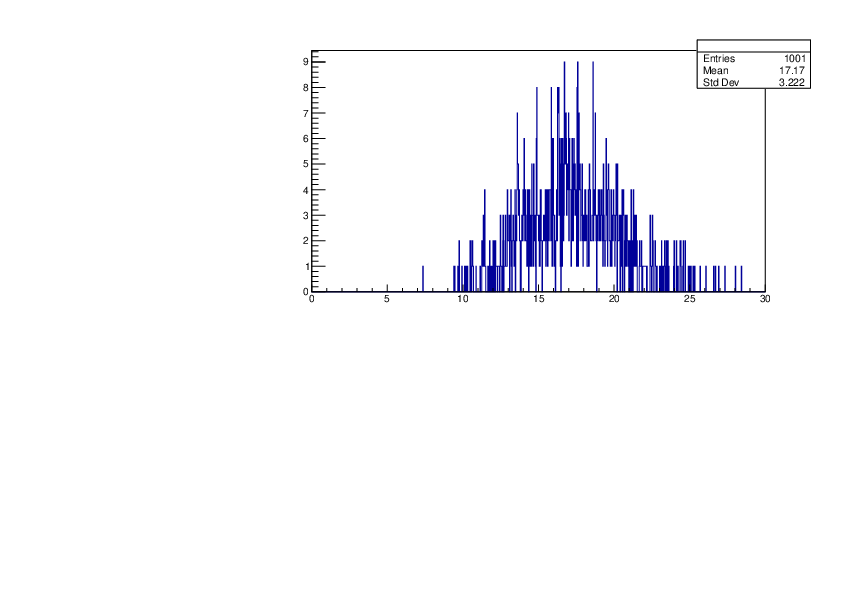
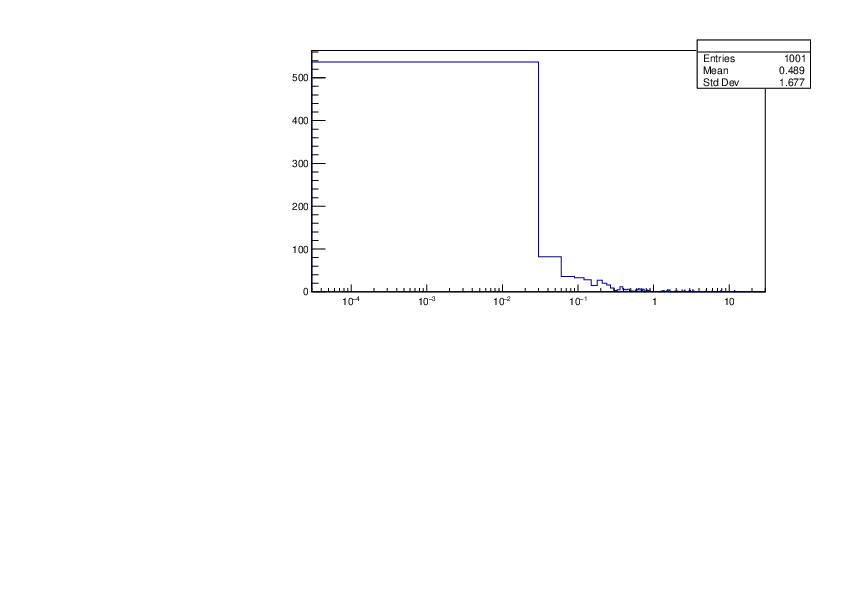
Selin Ezgi Birincioglu | Ca42+n Variations By Different Models |
The variations of MACS distribution of Ca42+n with different nuclear models generated by TALYS. When the level density is high enough, the resonances seem to decrease as TALYS obeys the statistical model. |
| Attachment 1: ld1.pdf
|
|
| Attachment 2: ld2.pdf
|
|
| Attachment 3: ld3.pdf
|
|
| Attachment 4: ld4.pdf
|
|
| Attachment 5: ld5.pdf
|
|
| Attachment 6: ld6.pdf
|
|
|
13
|
Thu Nov 30 13:54:25 2023 |
Selin Ezgi Birincioglu | Files for Na23+n |
1 -> macs.root: MACS distribution file
2 -> read.C: file to graph the histogram
3 -> list.dat: reads every run done by mygoe.f
4 -> talys.input: TALYS inputs
5 -> talys.output: output parameters for mygoe.f
6 -> Na24run: to run mygoe.f for Na23+n parameters taken from TALYS |
| Attachment 1: macs.root
|
| Attachment 2: read.C
|
#include "TRandom.h"
#include <TPolyLine.h>
#include <time.h>
#include "TTree.h"
#include <TROOT.h>
#include <TApplication.h>
#include <TRint.h>
#include <TSystem.h>
#include <TH1.h>
#include <TH2.h>
#include <TAxis.h>
#include <TGaxis.h>
#include <TCanvas.h>
#include <TGraph.h>
#include <TGraphErrors.h>
#include <TGraphAsymmErrors.h>
#include <TMultiGraph.h>
#include <TStyle.h>
#include <TKey.h>
#include <TLegend.h>
#include <TColor.h>
#include <TPad.h>
#include <TText.h>
#include <TPaveText.h>
#include <TBox.h>
#include <TLine.h>
#include <TMarker.h>
#include <TLatex.h>
#include <TMath.h>
#include <TF1.h>
#include <TFile.h>
#include <TClass.h>
#include "TBrowser.h"
#include "TFrame.h"
#include "TLegendEntry.h"
#include "TPave.h"
#include <fstream>
#include "Math/Minimizer.h"
#include "Math/Factory.h"
#include "Math/Functor.h"
#include <TVirtualFitter.h>
using namespace std;
float massAl=23.;
float massn=1.008665;
float massfac=massAl/(massAl+massn);
//constants
float kT=20000;
const double hbar=1.05457182*pow(10,-34);
const double kb=1.380649*pow(10,-23);
const double eVtoK=11606; // 1 eV is 11606 K
const double eVtojoule=1.60218*pow(10,-19);
const double pi=3.14159265359;
const double amu=1.66054*pow(10,-27);
double mu=massfac*amu;
const double Temp=kT*eVtoK;
const double vT=sqrt(2*kb*Temp/mu);
Double_t getmacs(string filename);
void read(){
TH1F *hmacs=new TH1F("","",5000,0,4);
TH1F *hmacs0=new TH1F("","",5000,0,4);
TH1F *hmacs1=new TH1F("","",5000,0,4);
ifstream list1;
list1.open("list.dat");
string fname1;
string fname2;
string fname3;
string fname4;
string fname5;
string fname6;
if(list1.good()){
while(1){
if(!list1.good())break;
list1>>fname1>>fname2>>fname3>>fname4>>fname5>>fname6;
cout<<fname1<<" "<<fname2<<endl;
Double_t macs0=getmacs(fname1);
Double_t macs02=getmacs(fname2);
Double_t macs1=getmacs(fname3);
Double_t macs12=getmacs(fname4);
Double_t macs13=getmacs(fname5);
Double_t macs14=getmacs(fname6);
double swave=macs0+macs02;
double pwave=macs1+macs12+macs13+macs14;
cout<<fname1<<" "<<swave<<" "<<pwave<<" "<<swave+pwave<<endl;
double total=1000*(swave+pwave);
hmacs->Fill(total);
hmacs0->Fill(swave*1000);
hmacs1->Fill(pwave*1000);
}
list1.close();
}
TFile *f=new TFile("macs.root","recreate");
hmacs->Write("macs");
hmacs0->Write("macs0");
hmacs1->Write("macs1");
f->Write();
}
Double_t getmacs(string filename){
int number, numberold;
numberold=0;
float sum =0;
//TString name1;
Double_t energy, spin, ell, wtot, wn,wg,wx, gw, kernel;
//string name1=filename;
ifstream finp;
finp.open(filename);
//cout<<filename<<endl;
for(int i=0;i<28;i++){
string line; getline(finp,line);}
string line;
while(1){
if(!finp)break;
finp>>number>>energy>>spin>>ell>>wtot>>wn>>wg>>wx>>gw>>kernel;
if(number>numberold){
sum=sum+kernel*exp(-energy/kT);
//cout<<number<< " "<<energy<<" "<<sum<<" "<<kernel<<endl;
number=numberold;
}
}
//cout<<mu<<" "<<kb<<" "<<Temp<<" "<<vT<<endl;
double macs=pow(2*pi/mu/kb/Temp,1.5)*hbar*hbar*eVtojoule*sum*1E28/vT;
//cout<<"MACS = "<<macs<<" b"<<endl;
finp.close();
return macs;
}
void test(){
int number, numberold;
numberold=0;
float sum =0;
//TString name1;
Double_t energy, spin, ell, wtot, wn,wg,wx, gw, kernel;
//string name1=filename;
ifstream finp;
finp.open("run1_25m_0l_1");
for(int i=0;i<28;i++){
string line; getline(finp,line);}
string line;
while(1){
if(!finp)break;
finp>>number>>energy>>spin>>ell>>wtot>>wn>>wg>>wx>>gw>>kernel;
if(number>numberold){
sum=sum+kernel*exp(-energy/kT);
cout<<number<< " "<<energy<<" "<<sum<<" "<<kernel<<endl;
number=numberold;
}
}
//cout<<mu<<" "<<kb<<" "<<Temp<<" "<<vT<<endl;
double macs=pow(2*pi/mu/kb/Temp,1.5)*hbar*hbar*eVtojoule*sum*1E28/vT;
cout<<"MACS = "<<macs<<" b"<<endl;
finp.close();
}
|
| Attachment 3: list.dat
|
run1_0l_1 run2_0l_1 run1_1l_1 run2_1l_1 run3_1l_1 run4_1l_1
run1_0l_10 run2_0l_10 run1_1l_10 run2_1l_10 run3_1l_10 run4_1l_10
run1_0l_100 run2_0l_100 run1_1l_100 run2_1l_100 run3_1l_100 run4_1l_100
run1_0l_1000 run2_0l_1000 run1_1l_1000 run2_1l_1000 run3_1l_1000 run4_1l_1000
run1_0l_101 run2_0l_101 run1_1l_101 run2_1l_101 run3_1l_101 run4_1l_101
run1_0l_102 run2_0l_102 run1_1l_102 run2_1l_102 run3_1l_102 run4_1l_102
run1_0l_103 run2_0l_103 run1_1l_103 run2_1l_103 run3_1l_103 run4_1l_103
run1_0l_104 run2_0l_104 run1_1l_104 run2_1l_104 run3_1l_104 run4_1l_104
run1_0l_105 run2_0l_105 run1_1l_105 run2_1l_105 run3_1l_105 run4_1l_105
run1_0l_106 run2_0l_106 run1_1l_106 run2_1l_106 run3_1l_106 run4_1l_106
run1_0l_107 run2_0l_107 run1_1l_107 run2_1l_107 run3_1l_107 run4_1l_107
run1_0l_108 run2_0l_108 run1_1l_108 run2_1l_108 run3_1l_108 run4_1l_108
run1_0l_109 run2_0l_109 run1_1l_109 run2_1l_109 run3_1l_109 run4_1l_109
run1_0l_11 run2_0l_11 run1_1l_11 run2_1l_11 run3_1l_11 run4_1l_11
run1_0l_110 run2_0l_110 run1_1l_110 run2_1l_110 run3_1l_110 run4_1l_110
run1_0l_111 run2_0l_111 run1_1l_111 run2_1l_111 run3_1l_111 run4_1l_111
run1_0l_112 run2_0l_112 run1_1l_112 run2_1l_112 run3_1l_112 run4_1l_112
run1_0l_113 run2_0l_113 run1_1l_113 run2_1l_113 run3_1l_113 run4_1l_113
run1_0l_114 run2_0l_114 run1_1l_114 run2_1l_114 run3_1l_114 run4_1l_114
run1_0l_115 run2_0l_115 run1_1l_115 run2_1l_115 run3_1l_115 run4_1l_115
run1_0l_116 run2_0l_116 run1_1l_116 run2_1l_116 run3_1l_116 run4_1l_116
run1_0l_117 run2_0l_117 run1_1l_117 run2_1l_117 run3_1l_117 run4_1l_117
run1_0l_118 run2_0l_118 run1_1l_118 run2_1l_118 run3_1l_118 run4_1l_118
run1_0l_119 run2_0l_119 run1_1l_119 run2_1l_119 run3_1l_119 run4_1l_119
run1_0l_12 run2_0l_12 run1_1l_12 run2_1l_12 run3_1l_12 run4_1l_12
run1_0l_120 run2_0l_120 run1_1l_120 run2_1l_120 run3_1l_120 run4_1l_120
run1_0l_121 run2_0l_121 run1_1l_121 run2_1l_121 run3_1l_121 run4_1l_121
run1_0l_122 run2_0l_122 run1_1l_122 run2_1l_122 run3_1l_122 run4_1l_122
run1_0l_123 run2_0l_123 run1_1l_123 run2_1l_123 run3_1l_123 run4_1l_123
run1_0l_124 run2_0l_124 run1_1l_124 run2_1l_124 run3_1l_124 run4_1l_124
run1_0l_125 run2_0l_125 run1_1l_125 run2_1l_125 run3_1l_125 run4_1l_125
run1_0l_126 run2_0l_126 run1_1l_126 run2_1l_126 run3_1l_126 run4_1l_126
run1_0l_127 run2_0l_127 run1_1l_127 run2_1l_127 run3_1l_127 run4_1l_127
run1_0l_128 run2_0l_128 run1_1l_128 run2_1l_128 run3_1l_128 run4_1l_128
run1_0l_129 run2_0l_129 run1_1l_129 run2_1l_129 run3_1l_129 run4_1l_129
run1_0l_13 run2_0l_13 run1_1l_13 run2_1l_13 run3_1l_13 run4_1l_13
run1_0l_130 run2_0l_130 run1_1l_130 run2_1l_130 run3_1l_130 run4_1l_130
run1_0l_131 run2_0l_131 run1_1l_131 run2_1l_131 run3_1l_131 run4_1l_131
run1_0l_132 run2_0l_132 run1_1l_132 run2_1l_132 run3_1l_132 run4_1l_132
run1_0l_133 run2_0l_133 run1_1l_133 run2_1l_133 run3_1l_133 run4_1l_133
run1_0l_134 run2_0l_134 run1_1l_134 run2_1l_134 run3_1l_134 run4_1l_134
run1_0l_135 run2_0l_135 run1_1l_135 run2_1l_135 run3_1l_135 run4_1l_135
run1_0l_136 run2_0l_136 run1_1l_136 run2_1l_136 run3_1l_136 run4_1l_136
run1_0l_137 run2_0l_137 run1_1l_137 run2_1l_137 run3_1l_137 run4_1l_137
run1_0l_138 run2_0l_138 run1_1l_138 run2_1l_138 run3_1l_138 run4_1l_138
run1_0l_139 run2_0l_139 run1_1l_139 run2_1l_139 run3_1l_139 run4_1l_139
run1_0l_14 run2_0l_14 run1_1l_14 run2_1l_14 run3_1l_14 run4_1l_14
run1_0l_140 run2_0l_140 run1_1l_140 run2_1l_140 run3_1l_140 run4_1l_140
run1_0l_141 run2_0l_141 run1_1l_141 run2_1l_141 run3_1l_141 run4_1l_141
run1_0l_142 run2_0l_142 run1_1l_142 run2_1l_142 run3_1l_142 run4_1l_142
run1_0l_143 run2_0l_143 run1_1l_143 run2_1l_143 run3_1l_143 run4_1l_143
run1_0l_144 run2_0l_144 run1_1l_144 run2_1l_144 run3_1l_144 run4_1l_144
run1_0l_145 run2_0l_145 run1_1l_145 run2_1l_145 run3_1l_145 run4_1l_145
run1_0l_146 run2_0l_146 run1_1l_146 run2_1l_146 run3_1l_146 run4_1l_146
run1_0l_147 run2_0l_147 run1_1l_147 run2_1l_147 run3_1l_147 run4_1l_147
run1_0l_148 run2_0l_148 run1_1l_148 run2_1l_148 run3_1l_148 run4_1l_148
run1_0l_149 run2_0l_149 run1_1l_149 run2_1l_149 run3_1l_149 run4_1l_149
run1_0l_15 run2_0l_15 run1_1l_15 run2_1l_15 run3_1l_15 run4_1l_15
run1_0l_150 run2_0l_150 run1_1l_150 run2_1l_150 run3_1l_150 run4_1l_150
run1_0l_151 run2_0l_151 run1_1l_151 run2_1l_151 run3_1l_151 run4_1l_151
run1_0l_152 run2_0l_152 run1_1l_152 run2_1l_152 run3_1l_152 run4_1l_152
run1_0l_153 run2_0l_153 run1_1l_153 run2_1l_153 run3_1l_153 run4_1l_153
run1_0l_154 run2_0l_154 run1_1l_154 run2_1l_154 run3_1l_154 run4_1l_154
run1_0l_155 run2_0l_155 run1_1l_155 run2_1l_155 run3_1l_155 run4_1l_155
run1_0l_156 run2_0l_156 run1_1l_156 run2_1l_156 run3_1l_156 run4_1l_156
run1_0l_157 run2_0l_157 run1_1l_157 run2_1l_157 run3_1l_157 run4_1l_157
run1_0l_158 run2_0l_158 run1_1l_158 run2_1l_158 run3_1l_158 run4_1l_158
run1_0l_159 run2_0l_159 run1_1l_159 run2_1l_159 run3_1l_159 run4_1l_159
run1_0l_16 run2_0l_16 run1_1l_16 run2_1l_16 run3_1l_16 run4_1l_16
run1_0l_160 run2_0l_160 run1_1l_160 run2_1l_160 run3_1l_160 run4_1l_160
run1_0l_161 run2_0l_161 run1_1l_161 run2_1l_161 run3_1l_161 run4_1l_161
run1_0l_162 run2_0l_162 run1_1l_162 run2_1l_162 run3_1l_162 run4_1l_162
run1_0l_163 run2_0l_163 run1_1l_163 run2_1l_163 run3_1l_163 run4_1l_163
run1_0l_164 run2_0l_164 run1_1l_164 run2_1l_164 run3_1l_164 run4_1l_164
run1_0l_165 run2_0l_165 run1_1l_165 run2_1l_165 run3_1l_165 run4_1l_165
run1_0l_166 run2_0l_166 run1_1l_166 run2_1l_166 run3_1l_166 run4_1l_166
run1_0l_167 run2_0l_167 run1_1l_167 run2_1l_167 run3_1l_167 run4_1l_167
run1_0l_168 run2_0l_168 run1_1l_168 run2_1l_168 run3_1l_168 run4_1l_168
run1_0l_169 run2_0l_169 run1_1l_169 run2_1l_169 run3_1l_169 run4_1l_169
run1_0l_17 run2_0l_17 run1_1l_17 run2_1l_17 run3_1l_17 run4_1l_17
run1_0l_170 run2_0l_170 run1_1l_170 run2_1l_170 run3_1l_170 run4_1l_170
run1_0l_171 run2_0l_171 run1_1l_171 run2_1l_171 run3_1l_171 run4_1l_171
run1_0l_172 run2_0l_172 run1_1l_172 run2_1l_172 run3_1l_172 run4_1l_172
run1_0l_173 run2_0l_173 run1_1l_173 run2_1l_173 run3_1l_173 run4_1l_173
run1_0l_174 run2_0l_174 run1_1l_174 run2_1l_174 run3_1l_174 run4_1l_174
run1_0l_175 run2_0l_175 run1_1l_175 run2_1l_175 run3_1l_175 run4_1l_175
run1_0l_176 run2_0l_176 run1_1l_176 run2_1l_176 run3_1l_176 run4_1l_176
run1_0l_177 run2_0l_177 run1_1l_177 run2_1l_177 run3_1l_177 run4_1l_177
run1_0l_178 run2_0l_178 run1_1l_178 run2_1l_178 run3_1l_178 run4_1l_178
run1_0l_179 run2_0l_179 run1_1l_179 run2_1l_179 run3_1l_179 run4_1l_179
run1_0l_18 run2_0l_18 run1_1l_18 run2_1l_18 run3_1l_18 run4_1l_18
run1_0l_180 run2_0l_180 run1_1l_180 run2_1l_180 run3_1l_180 run4_1l_180
run1_0l_181 run2_0l_181 run1_1l_181 run2_1l_181 run3_1l_181 run4_1l_181
run1_0l_182 run2_0l_182 run1_1l_182 run2_1l_182 run3_1l_182 run4_1l_182
run1_0l_183 run2_0l_183 run1_1l_183 run2_1l_183 run3_1l_183 run4_1l_183
run1_0l_184 run2_0l_184 run1_1l_184 run2_1l_184 run3_1l_184 run4_1l_184
run1_0l_185 run2_0l_185 run1_1l_185 run2_1l_185 run3_1l_185 run4_1l_185
run1_0l_186 run2_0l_186 run1_1l_186 run2_1l_186 run3_1l_186 run4_1l_186
run1_0l_187 run2_0l_187 run1_1l_187 run2_1l_187 run3_1l_187 run4_1l_187
run1_0l_188 run2_0l_188 run1_1l_188 run2_1l_188 run3_1l_188 run4_1l_188
run1_0l_189 run2_0l_189 run1_1l_189 run2_1l_189 run3_1l_189 run4_1l_189
run1_0l_19 run2_0l_19 run1_1l_19 run2_1l_19 run3_1l_19 run4_1l_19
run1_0l_190 run2_0l_190 run1_1l_190 run2_1l_190 run3_1l_190 run4_1l_190
run1_0l_191 run2_0l_191 run1_1l_191 run2_1l_191 run3_1l_191 run4_1l_191
run1_0l_192 run2_0l_192 run1_1l_192 run2_1l_192 run3_1l_192 run4_1l_192
run1_0l_193 run2_0l_193 run1_1l_193 run2_1l_193 run3_1l_193 run4_1l_193
run1_0l_194 run2_0l_194 run1_1l_194 run2_1l_194 run3_1l_194 run4_1l_194
run1_0l_195 run2_0l_195 run1_1l_195 run2_1l_195 run3_1l_195 run4_1l_195
run1_0l_196 run2_0l_196 run1_1l_196 run2_1l_196 run3_1l_196 run4_1l_196
run1_0l_197 run2_0l_197 run1_1l_197 run2_1l_197 run3_1l_197 run4_1l_197
run1_0l_198 run2_0l_198 run1_1l_198 run2_1l_198 run3_1l_198 run4_1l_198
run1_0l_199 run2_0l_199 run1_1l_199 run2_1l_199 run3_1l_199 run4_1l_199
run1_0l_2 run2_0l_2 run1_1l_2 run2_1l_2 run3_1l_2 run4_1l_2
run1_0l_20 run2_0l_20 run1_1l_20 run2_1l_20 run3_1l_20 run4_1l_20
run1_0l_200 run2_0l_200 run1_1l_200 run2_1l_200 run3_1l_200 run4_1l_200
run1_0l_201 run2_0l_201 run1_1l_201 run2_1l_201 run3_1l_201 run4_1l_201
run1_0l_202 run2_0l_202 run1_1l_202 run2_1l_202 run3_1l_202 run4_1l_202
run1_0l_203 run2_0l_203 run1_1l_203 run2_1l_203 run3_1l_203 run4_1l_203
run1_0l_204 run2_0l_204 run1_1l_204 run2_1l_204 run3_1l_204 run4_1l_204
run1_0l_205 run2_0l_205 run1_1l_205 run2_1l_205 run3_1l_205 run4_1l_205
run1_0l_206 run2_0l_206 run1_1l_206 run2_1l_206 run3_1l_206 run4_1l_206
run1_0l_207 run2_0l_207 run1_1l_207 run2_1l_207 run3_1l_207 run4_1l_207
run1_0l_208 run2_0l_208 run1_1l_208 run2_1l_208 run3_1l_208 run4_1l_208
run1_0l_209 run2_0l_209 run1_1l_209 run2_1l_209 run3_1l_209 run4_1l_209
run1_0l_21 run2_0l_21 run1_1l_21 run2_1l_21 run3_1l_21 run4_1l_21
run1_0l_210 run2_0l_210 run1_1l_210 run2_1l_210 run3_1l_210 run4_1l_210
run1_0l_211 run2_0l_211 run1_1l_211 run2_1l_211 run3_1l_211 run4_1l_211
run1_0l_212 run2_0l_212 run1_1l_212 run2_1l_212 run3_1l_212 run4_1l_212
run1_0l_213 run2_0l_213 run1_1l_213 run2_1l_213 run3_1l_213 run4_1l_213
run1_0l_214 run2_0l_214 run1_1l_214 run2_1l_214 run3_1l_214 run4_1l_214
run1_0l_215 run2_0l_215 run1_1l_215 run2_1l_215 run3_1l_215 run4_1l_215
run1_0l_216 run2_0l_216 run1_1l_216 run2_1l_216 run3_1l_216 run4_1l_216
run1_0l_217 run2_0l_217 run1_1l_217 run2_1l_217 run3_1l_217 run4_1l_217
run1_0l_218 run2_0l_218 run1_1l_218 run2_1l_218 run3_1l_218 run4_1l_218
run1_0l_219 run2_0l_219 run1_1l_219 run2_1l_219 run3_1l_219 run4_1l_219
run1_0l_22 run2_0l_22 run1_1l_22 run2_1l_22 run3_1l_22 run4_1l_22
run1_0l_220 run2_0l_220 run1_1l_220 run2_1l_220 run3_1l_220 run4_1l_220
run1_0l_221 run2_0l_221 run1_1l_221 run2_1l_221 run3_1l_221 run4_1l_221
run1_0l_222 run2_0l_222 run1_1l_222 run2_1l_222 run3_1l_222 run4_1l_222
run1_0l_223 run2_0l_223 run1_1l_223 run2_1l_223 run3_1l_223 run4_1l_223
run1_0l_224 run2_0l_224 run1_1l_224 run2_1l_224 run3_1l_224 run4_1l_224
run1_0l_225 run2_0l_225 run1_1l_225 run2_1l_225 run3_1l_225 run4_1l_225
run1_0l_226 run2_0l_226 run1_1l_226 run2_1l_226 run3_1l_226 run4_1l_226
run1_0l_227 run2_0l_227 run1_1l_227 run2_1l_227 run3_1l_227 run4_1l_227
run1_0l_228 run2_0l_228 run1_1l_228 run2_1l_228 run3_1l_228 run4_1l_228
run1_0l_229 run2_0l_229 run1_1l_229 run2_1l_229 run3_1l_229 run4_1l_229
run1_0l_23 run2_0l_23 run1_1l_23 run2_1l_23 run3_1l_23 run4_1l_23
run1_0l_230 run2_0l_230 run1_1l_230 run2_1l_230 run3_1l_230 run4_1l_230
run1_0l_231 run2_0l_231 run1_1l_231 run2_1l_231 run3_1l_231 run4_1l_231
run1_0l_232 run2_0l_232 run1_1l_232 run2_1l_232 run3_1l_232 run4_1l_232
run1_0l_233 run2_0l_233 run1_1l_233 run2_1l_233 run3_1l_233 run4_1l_233
run1_0l_234 run2_0l_234 run1_1l_234 run2_1l_234 run3_1l_234 run4_1l_234
run1_0l_235 run2_0l_235 run1_1l_235 run2_1l_235 run3_1l_235 run4_1l_235
run1_0l_236 run2_0l_236 run1_1l_236 run2_1l_236 run3_1l_236 run4_1l_236
run1_0l_237 run2_0l_237 run1_1l_237 run2_1l_237 run3_1l_237 run4_1l_237
run1_0l_238 run2_0l_238 run1_1l_238 run2_1l_238 run3_1l_238 run4_1l_238
run1_0l_239 run2_0l_239 run1_1l_239 run2_1l_239 run3_1l_239 run4_1l_239
run1_0l_24 run2_0l_24 run1_1l_24 run2_1l_24 run3_1l_24 run4_1l_24
run1_0l_240 run2_0l_240 run1_1l_240 run2_1l_240 run3_1l_240 run4_1l_240
run1_0l_241 run2_0l_241 run1_1l_241 run2_1l_241 run3_1l_241 run4_1l_241
run1_0l_242 run2_0l_242 run1_1l_242 run2_1l_242 run3_1l_242 run4_1l_242
run1_0l_243 run2_0l_243 run1_1l_243 run2_1l_243 run3_1l_243 run4_1l_243
run1_0l_244 run2_0l_244 run1_1l_244 run2_1l_244 run3_1l_244 run4_1l_244
run1_0l_245 run2_0l_245 run1_1l_245 run2_1l_245 run3_1l_245 run4_1l_245
run1_0l_246 run2_0l_246 run1_1l_246 run2_1l_246 run3_1l_246 run4_1l_246
run1_0l_247 run2_0l_247 run1_1l_247 run2_1l_247 run3_1l_247 run4_1l_247
run1_0l_248 run2_0l_248 run1_1l_248 run2_1l_248 run3_1l_248 run4_1l_248
run1_0l_249 run2_0l_249 run1_1l_249 run2_1l_249 run3_1l_249 run4_1l_249
run1_0l_25 run2_0l_25 run1_1l_25 run2_1l_25 run3_1l_25 run4_1l_25
run1_0l_250 run2_0l_250 run1_1l_250 run2_1l_250 run3_1l_250 run4_1l_250
run1_0l_251 run2_0l_251 run1_1l_251 run2_1l_251 run3_1l_251 run4_1l_251
run1_0l_252 run2_0l_252 run1_1l_252 run2_1l_252 run3_1l_252 run4_1l_252
run1_0l_253 run2_0l_253 run1_1l_253 run2_1l_253 run3_1l_253 run4_1l_253
run1_0l_254 run2_0l_254 run1_1l_254 run2_1l_254 run3_1l_254 run4_1l_254
run1_0l_255 run2_0l_255 run1_1l_255 run2_1l_255 run3_1l_255 run4_1l_255
run1_0l_256 run2_0l_256 run1_1l_256 run2_1l_256 run3_1l_256 run4_1l_256
run1_0l_257 run2_0l_257 run1_1l_257 run2_1l_257 run3_1l_257 run4_1l_257
run1_0l_258 run2_0l_258 run1_1l_258 run2_1l_258 run3_1l_258 run4_1l_258
run1_0l_259 run2_0l_259 run1_1l_259 run2_1l_259 run3_1l_259 run4_1l_259
run1_0l_26 run2_0l_26 run1_1l_26 run2_1l_26 run3_1l_26 run4_1l_26
run1_0l_260 run2_0l_260 run1_1l_260 run2_1l_260 run3_1l_260 run4_1l_260
run1_0l_261 run2_0l_261 run1_1l_261 run2_1l_261 run3_1l_261 run4_1l_261
run1_0l_262 run2_0l_262 run1_1l_262 run2_1l_262 run3_1l_262 run4_1l_262
run1_0l_263 run2_0l_263 run1_1l_263 run2_1l_263 run3_1l_263 run4_1l_263
run1_0l_264 run2_0l_264 run1_1l_264 run2_1l_264 run3_1l_264 run4_1l_264
run1_0l_265 run2_0l_265 run1_1l_265 run2_1l_265 run3_1l_265 run4_1l_265
run1_0l_266 run2_0l_266 run1_1l_266 run2_1l_266 run3_1l_266 run4_1l_266
run1_0l_267 run2_0l_267 run1_1l_267 run2_1l_267 run3_1l_267 run4_1l_267
run1_0l_268 run2_0l_268 run1_1l_268 run2_1l_268 run3_1l_268 run4_1l_268
run1_0l_269 run2_0l_269 run1_1l_269 run2_1l_269 run3_1l_269 run4_1l_269
run1_0l_27 run2_0l_27 run1_1l_27 run2_1l_27 run3_1l_27 run4_1l_27
run1_0l_270 run2_0l_270 run1_1l_270 run2_1l_270 run3_1l_270 run4_1l_270
run1_0l_271 run2_0l_271 run1_1l_271 run2_1l_271 run3_1l_271 run4_1l_271
run1_0l_272 run2_0l_272 run1_1l_272 run2_1l_272 run3_1l_272 run4_1l_272
run1_0l_273 run2_0l_273 run1_1l_273 run2_1l_273 run3_1l_273 run4_1l_273
run1_0l_274 run2_0l_274 run1_1l_274 run2_1l_274 run3_1l_274 run4_1l_274
run1_0l_275 run2_0l_275 run1_1l_275 run2_1l_275 run3_1l_275 run4_1l_275
run1_0l_276 run2_0l_276 run1_1l_276 run2_1l_276 run3_1l_276 run4_1l_276
run1_0l_277 run2_0l_277 run1_1l_277 run2_1l_277 run3_1l_277 run4_1l_277
run1_0l_278 run2_0l_278 run1_1l_278 run2_1l_278 run3_1l_278 run4_1l_278
run1_0l_279 run2_0l_279 run1_1l_279 run2_1l_279 run3_1l_279 run4_1l_279
run1_0l_28 run2_0l_28 run1_1l_28 run2_1l_28 run3_1l_28 run4_1l_28
run1_0l_280 run2_0l_280 run1_1l_280 run2_1l_280 run3_1l_280 run4_1l_280
run1_0l_281 run2_0l_281 run1_1l_281 run2_1l_281 run3_1l_281 run4_1l_281
run1_0l_282 run2_0l_282 run1_1l_282 run2_1l_282 run3_1l_282 run4_1l_282
run1_0l_283 run2_0l_283 run1_1l_283 run2_1l_283 run3_1l_283 run4_1l_283
run1_0l_284 run2_0l_284 run1_1l_284 run2_1l_284 run3_1l_284 run4_1l_284
run1_0l_285 run2_0l_285 run1_1l_285 run2_1l_285 run3_1l_285 run4_1l_285
run1_0l_286 run2_0l_286 run1_1l_286 run2_1l_286 run3_1l_286 run4_1l_286
run1_0l_287 run2_0l_287 run1_1l_287 run2_1l_287 run3_1l_287 run4_1l_287
run1_0l_288 run2_0l_288 run1_1l_288 run2_1l_288 run3_1l_288 run4_1l_288
run1_0l_289 run2_0l_289 run1_1l_289 run2_1l_289 run3_1l_289 run4_1l_289
run1_0l_29 run2_0l_29 run1_1l_29 run2_1l_29 run3_1l_29 run4_1l_29
run1_0l_290 run2_0l_290 run1_1l_290 run2_1l_290 run3_1l_290 run4_1l_290
run1_0l_291 run2_0l_291 run1_1l_291 run2_1l_291 run3_1l_291 run4_1l_291
run1_0l_292 run2_0l_292 run1_1l_292 run2_1l_292 run3_1l_292 run4_1l_292
run1_0l_293 run2_0l_293 run1_1l_293 run2_1l_293 run3_1l_293 run4_1l_293
run1_0l_294 run2_0l_294 run1_1l_294 run2_1l_294 run3_1l_294 run4_1l_294
run1_0l_295 run2_0l_295 run1_1l_295 run2_1l_295 run3_1l_295 run4_1l_295
run1_0l_296 run2_0l_296 run1_1l_296 run2_1l_296 run3_1l_296 run4_1l_296
run1_0l_297 run2_0l_297 run1_1l_297 run2_1l_297 run3_1l_297 run4_1l_297
run1_0l_298 run2_0l_298 run1_1l_298 run2_1l_298 run3_1l_298 run4_1l_298
run1_0l_299 run2_0l_299 run1_1l_299 run2_1l_299 run3_1l_299 run4_1l_299
run1_0l_3 run2_0l_3 run1_1l_3 run2_1l_3 run3_1l_3 run4_1l_3
run1_0l_30 run2_0l_30 run1_1l_30 run2_1l_30 run3_1l_30 run4_1l_30
run1_0l_300 run2_0l_300 run1_1l_300 run2_1l_300 run3_1l_300 run4_1l_300
run1_0l_301 run2_0l_301 run1_1l_301 run2_1l_301 run3_1l_301 run4_1l_301
run1_0l_302 run2_0l_302 run1_1l_302 run2_1l_302 run3_1l_302 run4_1l_302
run1_0l_303 run2_0l_303 run1_1l_303 run2_1l_303 run3_1l_303 run4_1l_303
run1_0l_304 run2_0l_304 run1_1l_304 run2_1l_304 run3_1l_304 run4_1l_304
run1_0l_305 run2_0l_305 run1_1l_305 run2_1l_305 run3_1l_305 run4_1l_305
run1_0l_306 run2_0l_306 run1_1l_306 run2_1l_306 run3_1l_306 run4_1l_306
run1_0l_307 run2_0l_307 run1_1l_307 run2_1l_307 run3_1l_307 run4_1l_307
run1_0l_308 run2_0l_308 run1_1l_308 run2_1l_308 run3_1l_308 run4_1l_308
run1_0l_309 run2_0l_309 run1_1l_309 run2_1l_309 run3_1l_309 run4_1l_309
run1_0l_31 run2_0l_31 run1_1l_31 run2_1l_31 run3_1l_31 run4_1l_31
run1_0l_310 run2_0l_310 run1_1l_310 run2_1l_310 run3_1l_310 run4_1l_310
run1_0l_311 run2_0l_311 run1_1l_311 run2_1l_311 run3_1l_311 run4_1l_311
run1_0l_312 run2_0l_312 run1_1l_312 run2_1l_312 run3_1l_312 run4_1l_312
run1_0l_313 run2_0l_313 run1_1l_313 run2_1l_313 run3_1l_313 run4_1l_313
run1_0l_314 run2_0l_314 run1_1l_314 run2_1l_314 run3_1l_314 run4_1l_314
run1_0l_315 run2_0l_315 run1_1l_315 run2_1l_315 run3_1l_315 run4_1l_315
run1_0l_316 run2_0l_316 run1_1l_316 run2_1l_316 run3_1l_316 run4_1l_316
run1_0l_317 run2_0l_317 run1_1l_317 run2_1l_317 run3_1l_317 run4_1l_317
run1_0l_318 run2_0l_318 run1_1l_318 run2_1l_318 run3_1l_318 run4_1l_318
run1_0l_319 run2_0l_319 run1_1l_319 run2_1l_319 run3_1l_319 run4_1l_319
run1_0l_32 run2_0l_32 run1_1l_32 run2_1l_32 run3_1l_32 run4_1l_32
run1_0l_320 run2_0l_320 run1_1l_320 run2_1l_320 run3_1l_320 run4_1l_320
run1_0l_321 run2_0l_321 run1_1l_321 run2_1l_321 run3_1l_321 run4_1l_321
run1_0l_322 run2_0l_322 run1_1l_322 run2_1l_322 run3_1l_322 run4_1l_322
run1_0l_323 run2_0l_323 run1_1l_323 run2_1l_323 run3_1l_323 run4_1l_323
run1_0l_324 run2_0l_324 run1_1l_324 run2_1l_324 run3_1l_324 run4_1l_324
run1_0l_325 run2_0l_325 run1_1l_325 run2_1l_325 run3_1l_325 run4_1l_325
run1_0l_326 run2_0l_326 run1_1l_326 run2_1l_326 run3_1l_326 run4_1l_326
run1_0l_327 run2_0l_327 run1_1l_327 run2_1l_327 run3_1l_327 run4_1l_327
run1_0l_328 run2_0l_328 run1_1l_328 run2_1l_328 run3_1l_328 run4_1l_328
run1_0l_329 run2_0l_329 run1_1l_329 run2_1l_329 run3_1l_329 run4_1l_329
run1_0l_33 run2_0l_33 run1_1l_33 run2_1l_33 run3_1l_33 run4_1l_33
run1_0l_330 run2_0l_330 run1_1l_330 run2_1l_330 run3_1l_330 run4_1l_330
run1_0l_331 run2_0l_331 run1_1l_331 run2_1l_331 run3_1l_331 run4_1l_331
run1_0l_332 run2_0l_332 run1_1l_332 run2_1l_332 run3_1l_332 run4_1l_332
run1_0l_333 run2_0l_333 run1_1l_333 run2_1l_333 run3_1l_333 run4_1l_333
run1_0l_334 run2_0l_334 run1_1l_334 run2_1l_334 run3_1l_334 run4_1l_334
run1_0l_335 run2_0l_335 run1_1l_335 run2_1l_335 run3_1l_335 run4_1l_335
run1_0l_336 run2_0l_336 run1_1l_336 run2_1l_336 run3_1l_336 run4_1l_336
run1_0l_337 run2_0l_337 run1_1l_337 run2_1l_337 run3_1l_337 run4_1l_337
run1_0l_338 run2_0l_338 run1_1l_338 run2_1l_338 run3_1l_338 run4_1l_338
run1_0l_339 run2_0l_339 run1_1l_339 run2_1l_339 run3_1l_339 run4_1l_339
run1_0l_34 run2_0l_34 run1_1l_34 run2_1l_34 run3_1l_34 run4_1l_34
run1_0l_340 run2_0l_340 run1_1l_340 run2_1l_340 run3_1l_340 run4_1l_340
run1_0l_341 run2_0l_341 run1_1l_341 run2_1l_341 run3_1l_341 run4_1l_341
run1_0l_342 run2_0l_342 run1_1l_342 run2_1l_342 run3_1l_342 run4_1l_342
run1_0l_343 run2_0l_343 run1_1l_343 run2_1l_343 run3_1l_343 run4_1l_343
run1_0l_344 run2_0l_344 run1_1l_344 run2_1l_344 run3_1l_344 run4_1l_344
run1_0l_345 run2_0l_345 run1_1l_345 run2_1l_345 run3_1l_345 run4_1l_345
run1_0l_346 run2_0l_346 run1_1l_346 run2_1l_346 run3_1l_346 run4_1l_346
run1_0l_347 run2_0l_347 run1_1l_347 run2_1l_347 run3_1l_347 run4_1l_347
run1_0l_348 run2_0l_348 run1_1l_348 run2_1l_348 run3_1l_348 run4_1l_348
run1_0l_349 run2_0l_349 run1_1l_349 run2_1l_349 run3_1l_349 run4_1l_349
run1_0l_35 run2_0l_35 run1_1l_35 run2_1l_35 run3_1l_35 run4_1l_35
run1_0l_350 run2_0l_350 run1_1l_350 run2_1l_350 run3_1l_350 run4_1l_350
run1_0l_351 run2_0l_351 run1_1l_351 run2_1l_351 run3_1l_351 run4_1l_351
run1_0l_352 run2_0l_352 run1_1l_352 run2_1l_352 run3_1l_352 run4_1l_352
run1_0l_353 run2_0l_353 run1_1l_353 run2_1l_353 run3_1l_353 run4_1l_353
run1_0l_354 run2_0l_354 run1_1l_354 run2_1l_354 run3_1l_354 run4_1l_354
run1_0l_355 run2_0l_355 run1_1l_355 run2_1l_355 run3_1l_355 run4_1l_355
run1_0l_356 run2_0l_356 run1_1l_356 run2_1l_356 run3_1l_356 run4_1l_356
run1_0l_357 run2_0l_357 run1_1l_357 run2_1l_357 run3_1l_357 run4_1l_357
run1_0l_358 run2_0l_358 run1_1l_358 run2_1l_358 run3_1l_358 run4_1l_358
run1_0l_359 run2_0l_359 run1_1l_359 run2_1l_359 run3_1l_359 run4_1l_359
run1_0l_36 run2_0l_36 run1_1l_36 run2_1l_36 run3_1l_36 run4_1l_36
run1_0l_360 run2_0l_360 run1_1l_360 run2_1l_360 run3_1l_360 run4_1l_360
run1_0l_361 run2_0l_361 run1_1l_361 run2_1l_361 run3_1l_361 run4_1l_361
run1_0l_362 run2_0l_362 run1_1l_362 run2_1l_362 run3_1l_362 run4_1l_362
run1_0l_363 run2_0l_363 run1_1l_363 run2_1l_363 run3_1l_363 run4_1l_363
run1_0l_364 run2_0l_364 run1_1l_364 run2_1l_364 run3_1l_364 run4_1l_364
run1_0l_365 run2_0l_365 run1_1l_365 run2_1l_365 run3_1l_365 run4_1l_365
run1_0l_366 run2_0l_366 run1_1l_366 run2_1l_366 run3_1l_366 run4_1l_366
run1_0l_367 run2_0l_367 run1_1l_367 run2_1l_367 run3_1l_367 run4_1l_367
run1_0l_368 run2_0l_368 run1_1l_368 run2_1l_368 run3_1l_368 run4_1l_368
... 701 more lines ...
|
| Attachment 4: talys.inp
|
#
# General
#
projectile n
element Na
mass 23
energy 1.
astroE 0.02
racap y
resonance y
#
# Output
#
#channels y
outgamma y
outdensity y
outlevels y
outbasic y
|
| Attachment 5: talys.out
|
TALYS-1.96 (Version: December 30, 2021)
Copyright (C) 2021 A.J. Koning, S. Hilaire and S. Goriely
Dimensions - Cross sections: mb, Energies: MeV, Angles: degrees
########## USER INPUT ##########
USER INPUT FILE
#
# general
#
projectile n
element na
mass 23
energy 1.
astroe 0.02
racap y
resonance y
#
# output
#
#channels y
outgamma y
outdensity y
outlevels y
outbasic y
USER INPUT FILE + DEFAULTS
Keyword Value Variable Explanation
#
# Four main keywords
#
projectile n ptype0 type of incident particle
element Na Starget symbol of target nucleus
mass 23 mass mass number of target nucleus
energy energyfile file with incident energies
#
# Basic physical and numerical parameters
#
ejectiles g n p d t h a outtype outgoing particles
maxz 8 maxZ maximal number of protons away from the initial compound nucleus
maxn 10 maxN maximal number of neutrons away from the initial compound nucleus
bins 40 nbins number of continuum excitation energy bins
equidistant y flagequi flag to use equidistant excitation bins instead of logarithmic bins
equispec n flagequispec flag to use equidistant bins for emission spectra
popmev n flagpopmev flag to use initial population per MeV instead of histograms
segment 1 segment number of segments to divide emission energy grid
maxlevelstar 30 nlevmax maximum number of included discrete levels for target
maxlevelsres 10 nlevmaxres maximum number of included discrete levels for residual nucleus
maxlevelsbin g 10 nlevbin maximum number of included discrete levels for gamma channel
maxlevelsbin n 30 nlevbin maximum number of included discrete levels for neutron channel
maxlevelsbin p 10 nlevbin maximum number of included discrete levels for proton channel
maxlevelsbin d 5 nlevbin maximum number of included discrete levels for deuteron channel
maxlevelsbin t 5 nlevbin maximum number of included discrete levels for triton channel
maxlevelsbin h 5 nlevbin maximum number of included discrete levels for helium-3 channel
maxlevelsbin a 10 nlevbin maximum number of included discrete levels for alpha channel
ltarget 0 ltarget excited level of target
isomer 1.00E+00 isomer definition of isomer in seconds
transpower 15 transpower power for transmission coefficient limit
transeps 1.00E-18 transeps limit for transmission coefficient
xseps 1.00E-17 xseps limit for cross sections
popeps 1.00E-13 popeps limit for population cross section per nucleus
Rfiseps 1.00E-06 Rfiseps ratio for limit for fission cross section per nucleus
elow 1.00E-06 elow minimal incident energy for nuclear model calculations
angles 90 nangle number of angles
anglescont 18 nanglecont number of angles for continuum
anglesrec 1 nanglerec number of recoil angles
maxenrec 20 maxenrec number of recoil energies
channels n flagchannels flag for exclusive channels calculation
maxchannel 4 maxchannel maximal number of outgoing particles in individual channel description
micro n flagmicro flag for completely microscopic Talys calculation
best n flagbest flag to use best set of adjusted parameters
bestbranch y flagbestbr flag to use flag to use only best set of branching ratios
bestend n flagbestend flag to put best set of parameters at end of input file
relativistic y flagrel flag for relativistic kinematics
recoil n flagrecoil flag for calculation of recoils
labddx n flaglabddx flag for calculation of DDX in LAB system
recoilaverage n flagrecoilav flag for average velocity in recoil calculation
channelenergy n flagEchannel flag for channel energy for emission spectrum
reaction y flagreaction flag for calculation of nuclear reactions
ngfit n flagngfit flag for using fitted (n,g) nuclear model parameters
nnfit n flagnnfit flag for using fitted (n,n), (n,2n) and (n,p) nuclear model parameters
nafit n flagnafit flag for using fitted (n,a) nuclear model parameters
astro y flagastro flag for calculation of astrophysics reaction rate
astrogs y flagastrogs flag for calculation of astrophysics reaction rate with target in ground state only
astroex n flagastroex flag for calculation of astrophysics reaction rate to long-lived excited states
nonthermlev -1 nonthermlev excited level non-thermalized in the calculation of astrophysics rate
massmodel 2 massmodel model for theoretical nuclear mass
expmass y flagexpmass flag for using experimental nuclear mass if available
disctable 1 disctable table with discrete levels
production n flagprod flag for isotope production
outfy n flagoutfy flag for output detailed fission yield calculation
gefran 50000 gefran number of random events for GEF calculation
Estop 1000.000 Estop incident energy above which TALYS stops
rpevap n flagrpevap flag for evaporation of residual products at high incident energies
maxZrp 12 maxZrp maximal number of protons away from the initial compound nucleus before residual evaporation
maxNrp 32 maxNrp maximal number of neutrons away from the initial compound nucleus before residual evaporation
#
# Optical model
#
localomp y flaglocalomp flag for local (y) or global (n) optical model
dispersion n flagdisp flag for dispersive optical model
jlmomp n flagjlm flag for using semi-microscopic JLM OMP
riplomp n flagriplomp flag for RIPL OMP
riplrisk n flagriplrisk flag for going outside RIPL mass validity range
optmodall n flagompall flag for new optical model calculation for all residual nuclei
incadjust y flagincadj flag for OMP adjustment on incident channel also
omponly n flagomponly flag to execute ONLY an optical model calculation
autorot n flagautorot flag for automatic rotational coupled channels calculations for A > 150
spherical n flagspher flag to force spherical optical model
soukho y flagsoukho flag for Soukhovitskii OMP for actinides
coulomb y flagcoulomb flag for Coulomb excitation calculation with ECIS
statepot n flagstate flag for optical model potential for each excited state
maxband 0 maxband highest vibrational band added to rotational model
maxrot 2 maxrot number of included excited rotational levels
sysreaction sysreaction particles with reaction cross section from systematics
rotational n rotational particles with possible rotational optical model
core -1 core even-even core for weakcoupling (-1 or 1)
ecissave n flagecissave flag for saving ECIS input and output files
eciscalc y flageciscalc flag for new ECIS calculation for outgoing particles and energy grid
inccalc y flaginccalc flag for new ECIS calculation for incident channel
endfecis y flagendfecis flag for new ECIS calculation for ENDF-6 files
radialmodel 2 radialmodel model for radial matter densities (JLM OMP only)
jlmmode 0 jlmmode option for JLM imaginary potential normalization
alphaomp 6 alphaomp alpha OMP (1=normal, 2= McFadden-Satchler, 3-5= folding potential, 6,8= Avrigeanu, 7=Nolte)
deuteronomp 1 deuteronomp deuteron OMP (1=normal, 2=Daehnick, 3=Bojowald, 4=Han-Shi-Shen, 5=An-Cai)
#
# Compound nucleus
#
widthfluc y flagwidth flag for width fluctuation calculation
widthmode 1 wmode designator for width fluctuation model
compound y flagcomp flag for compound nucleus model
fullhf n flagfullhf flag for full spin dependence of transmission coefficients
eciscompound n flageciscomp flag for compound nucleus calculation by ECIS
cpang n flagcpang flag for compound angular distribution calculation for incident charged particles
urr n flagurr flag for URR calculation
urrnjoy n flagurrnjoy flag for normalization of URR parameters with NJOY method
lurr 2 lurr maximal orbital angular momentum for URR
#
# Gamma emission
#
gammax 2 gammax number of l-values for gamma multipolarity
strength 9 strength model for E1 gamma-ray strength function
strengthM1 3 strengthM1 model for M1 gamma-ray strength function
electronconv y flagelectron flag for application of electron conversion coefficient
racap y flagracap flag for radiative capture model
ldmodelracap 1 ldmodelracap level density model for direct radiative capture
upbend y flagupbend flag for low-energy upbend of photon strength function
psfglobal n flagpsfglobal flag for global photon strength functions only
#
# Pre-equilibrium
#
preequilibrium n flagpreeq flag for pre-equilibrium calculation
preeqmode 2 preeqmode designator for pre-equilibrium model
multipreeq n flagmulpre flag for multiple pre-equilibrium calculation
mpreeqmode 2 mpreeqmode designator for multiple pre-equilibrium model
breakupmodel 1 breakupmodel model for break-up reaction: 1. Kalbach 2. Avrigeanu
phmodel 1 phmodel particle-hole state density model
pairmodel 2 pairmodel designator for pre-equilibrium pairing model
preeqspin 1 pespinmodel model for pre-equilibrium spin distribution
giantresonance y flaggiant flag for collective contribution from giant resonances
preeqsurface y flagsurface flag for surface effects in exciton model
preeqcomplex y flagpecomp flag for Kalbach complex particle emission model
twocomponent y flag2comp flag for two-component pre-equilibrium model
ecisdwba y flagecisdwba flag for new ECIS calculation for DWBA for MSD
onestep n flagonestep flag for continuum one-step direct only
#
# Level densities
#
ldmodel 1 ldmodelall level density model
ldmodelCN 1 ldmodelCN level density model for compound nucleus
shellmodel 1 shellmodel model for shell correction energies
kvibmodel 2 kvibmodel model for vibrational enhancement
spincutmodel 1 spincutmodel model for spin cutoff factor for ground state
asys n flagasys flag for all level density parameters a from systematics
parity n flagparity flag for non-equal parity distribution
colenhance n flagcolall flag for collective enhancement of level density for all nuclides
ctmglobal n flagctmglob flag for global CTM model (no discrete level info)
gshell n flaggshell flag for energy dependence of single particle level density parameter g
colldamp n flagcolldamp flag for damping of collective effects in effective level density
#
# Fission
#
fission n flagfission flag for fission
fismodel 3 fismodel fission model
fismodelalt 3 fismodelalt alternative fission model for default barriers
hbstate y flaghbstate flag for head band states in fission
class2 y flagclass2 flag for class2 states in fission
fispartdamp n flagfispartdamp flag for fission partial damping
massdis n flagmassdis flag for calculation of fission fragment mass yields
ffevaporation y flagffevap flag for calculation of particle evaporation from fission fragment mass yields
fisfeed n flagfisfeed flag for output of fission per excitation bin
fymodel 2 fymodel fission yield model, 1: Brosa 2: GEF
ffmodel 1 ffmodel fission fragment model, 1: GEF 2: HF3D (Okumura) 3: SPY
pfnsmodel 1 pfnsmodel PFNS model, 1: Iwamoto 2: from FF decay
ffspin n flagffspin flag to use spin distribution in initial FF population
#
# Output
#
outmain y flagmain flag for main output
outbasic y flagbasic flag for output of basic information and results
outpopulation y flagpop flag for output of population
outcheck y flagcheck flag for output of numerical checks
outlevels y flaglevels flag for output of discrete level information
outdensity y flagdensity flag for output of level densities
outomp y flagoutomp flag for output of optical model parameters
outkd n flagoutkd flag for output of KD03 OMP parameters
outdirect y flagdirect flag for output of direct reaction results
outinverse y flaginverse flag for output of transmission coefficients and inverse reaction cross sections
outdecay n flagdecay flag for output of decay of each population bin
outtransenergy y flagtransen flag for output of transmission coefficients per energy
outecis n flagoutecis flag for output of ECIS results
outgamma y flaggamma flag for output of gamma-ray information
outpreequilibrium n flagpeout flag for output of pre-equilibrium results
outfission n flagfisout flag for output of fission information
outdiscrete y flagdisc flag for output of discrete state cross sections
outspectra n flagspec flag for output of double-differential cross sections
outbinspectra n flagbinspec flag for output of emission spectrum per excitation bin
resonance y flagres flag for output of low energy resonance cross sections
group n flaggroup flag for output of low energy groupwise cross sections
addiscrete y flagadd flag for addition of discrete states to spectra
addelastic y flagaddel flag for addition of elastic peak to spectra
outangle n flagang flag for output of angular distributions
outlegendre n flaglegendre flag for output of Legendre coefficients
ddxmode 0 ddxmode mode for double-differential cross sections
outdwba n flagoutdwba flag for output of DWBA cross sections for MSD
outgamdis n flaggamdis flag for output of discrete gamma-ray intensities
outexcitation n flagexc flag for output of excitation functions
components n flagcompo flag for output of cross section components
endf n flagendf flag for information for ENDF-6 file
endfdetail y flagendfdet flag for detailed ENDF-6 information per channel
sacs n flagsacs flag for statistical analysis of cross sections
partable n flagpartable flag for output of model parameters on separate file
block n flagblock flag to block spectra, angle and gamma files
########## BASIC REACTION PARAMETERS ##########
Projectile : neutron Mass in a.m.u. : 1.008665
Target : 23Na Mass in a.m.u. : 22.989769
Included channels:
gamma
neutron
proton
deuteron
triton
helium-3
alpha
73 incident energies (LAB):
2.088E-06
4.175E-06
6.263E-06
8.351E-06
1.044E-05
2.088E-05
3.132E-05
4.175E-05
5.219E-05
6.263E-05
7.307E-05
8.351E-05
9.395E-05
1.044E-04
2.088E-04
3.132E-04
4.175E-04
5.219E-04
6.263E-04
7.307E-04
8.351E-04
9.395E-04
0.001
0.002
0.003
0.004
0.005
0.006
0.007
0.008
0.009
0.010
0.015
0.019
0.023
0.027
0.031
0.035
0.040
0.044
0.048
0.052
0.056
0.061
... 41556 more lines ...
|
| Attachment 6: Na24run
|
#!/bin/bash
rm -r Na24/
mkdir Na24
for((c=1;c<=1000;c++))
do
./a.out << EOF
23 1.5 # amass spin
0 1000000 #min max
0 98587.10938 0.7616e-4 #ell D S
0.47192 0.094384 #<G> SigmaG
0 0 #Gx sigma
0 #bias
EOF
cp out_all1 Na24/run1_0l_$c
cp out_all2 Na24/run2_0l_$c
rm out*
done
for((c=1;c<=1000;c++))
do
./a.out << EOF
23 1.5 # amass spin
0 1000000 #min max
1 63257.56250 3.1594e-4 #ell D S
0.92252 0.184504 #<G> SigmaG
0 0 #Gx sigma
0 #bias
EOF
cp out_all1 Na24/run1_1l_$c
cp out_all2 Na24/run2_1l_$c
cp out_all3 Na24/run3_1l_$c
cp out_all4 Na24/run4_1l_$c
rm out*
done
|